Autor: Sodré G. B. Neto
Afiliação: IPPTM – Instituto de Pesquisa em Paleogenética, TP53 e MicroRNA / CEGH / ICB / UFG
Resumo Estruturado
Introdução: A teoria da evolução biológica convencional fundamenta-se na premissa de que mutações aleatórias e seleção natural são capazes de gerar complexidade e viabilidade a longo prazo. No entanto, avanços na genética de populações e genômica comparativa sugerem um processo inverso: a degeneração sistemática do genoma pelo acúmulo de mutações deletérias, um fenômeno descrito como entropia genética [299].
Objetivo: Este artigo sintetiza evidências sobre o acúmulo de carga genética deletéria na linhagem humana e animal, propondo a ocorrência de um pico mutacional/radiativo recente durante o Holoceno, desencadeado por eventos catastróficos.
Métodos: Realizou-se uma revisão sistemática de literatura focada em taxas de mutação mitocondrial (mtDNA), acúmulo de mutações deletérias em populações pequenas e grandes, e o impacto de eventos catastróficos na geocronologia radioativa.
Resultados: As taxas de mutação observadas em estudos de linhagem (pedigree) são significativamente superiores às taxas filogenéticas tradicionais, indicando um erro sistemático na calibração do relógio molecular. Evidências geológicas sugerem que impactos de asteroides podem ter induzido picos de radiação via piezoeletricidade nuclear, acelerando o decaimento radioativo e as taxas mutacionais entre 5.000 e 10.000 anos atrás. A perda de funcionalidade em genes críticos, como o supressor tumoral p53, reflete essa senescência evolutiva.
Conclusão: A trajetória atual de acúmulo de mutações (deleteriome) aponta para um “mutational meltdown”, sugerindo que a viabilidade das espécies contemporâneas está severamente comprometida, com uma perspectiva de extinção iminente em termos geológicos.
Introdução
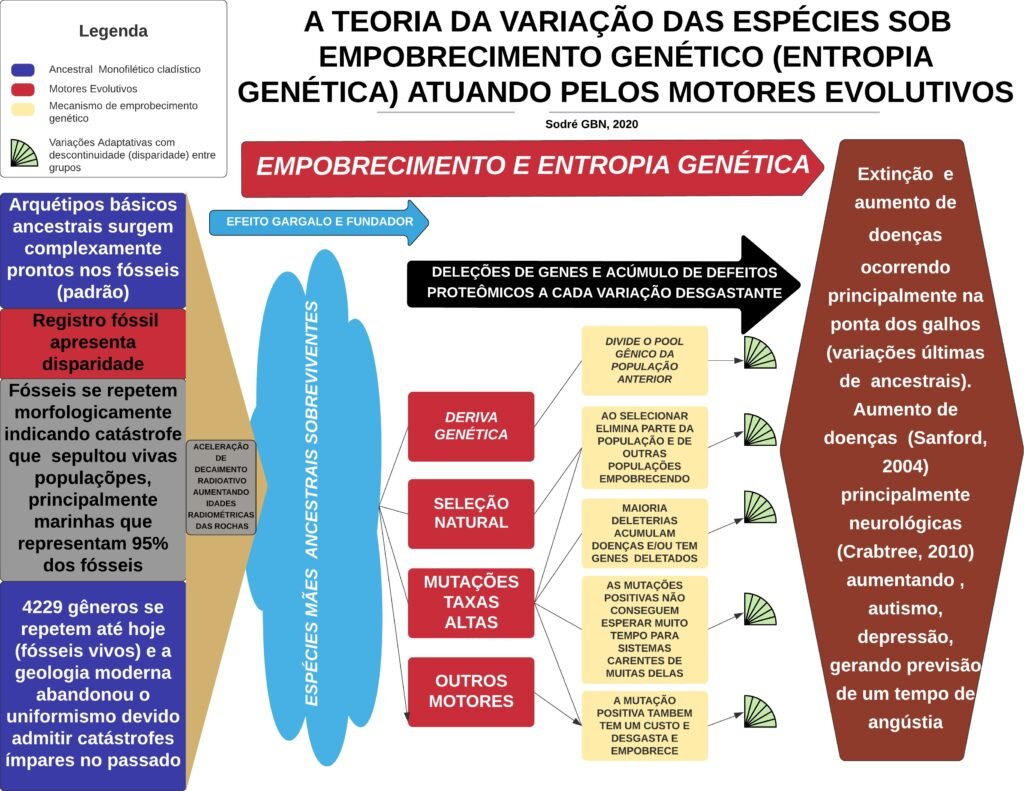
A narrativa evolutiva predominante postula que a vida na Terra se diversificou e se complexificou ao longo de bilhões de anos. Contudo, esta visão ignora um princípio fundamental da teoria da informação e da termodinâmica aplicado à biologia: a tendência natural ao desgaste e à perda de informação. A Teoria da Degeneração das Espécies (TDE) propõe que, longe de evoluir para formas mais complexas, os genomas estão em um estado de declínio progressivo devido ao acúmulo incessante de mutações deletérias.
O conceito de Entropia Genética, popularizado por John Sanford [299], argumenta que a grande maioria das mutações é “quase-neutra” e, portanto, invisível à seleção natural, acumulando-se silenciosamente em cada geração [8] [12] [259]. Este processo leva à degradação da aptidão biológica (fitness) e, eventualmente, ao colapso populacional [7] [13] [261]. Este artigo integra a TDE com evidências de um Pico Mutacional Holocênico, sugerindo que a história genética humana é muito mais curta e catastrófica do que o modelo uniformitarista sugere.
Métodos
A presente investigação baseia-se em uma meta-análise de dados genômicos e geocronológicos. Foram analisados:
1.Taxas de Mutação Mitocondrial: Comparação entre taxas baseadas em genealogia (pedigree) e taxas baseadas em calibrações fósseis [43] [45] [46] [47] [50] [52] [53] [56] [59] [60] [63] [65] [67] [68] [70] [71] [73] [74] [76] [77] [81].
2.Modelos de Acúmulo de Carga Genética: Avaliação do impacto de mutações deletérias na funcionalidade de proteínas essenciais [6] [7] [8] [9] [10] [11] [12] [13] [14] [15] [16] [17] [18] [19] [20] [21] [22] [23] [24] [25] [26] [27] [28] [29] [30] [31] [32] [33] [34] [35] [36] [37] [38] [39] [40] [41] [42] [48] [49] [51] [54] [55] [57] [58] [61] [62] [64] [66] [69] [72] [75] [78] [79] [80] [162] [163] [164] [165] [166] [167] [168] [169] [170] [171] [172] [173] [174] [175] [176] [177] [178] [179] [180] [181] [182] [183] [184] [185] [186] [187] [188] [189] [190] [191] [192] [193] [194] [195] [196] [197] [198] [199] [200] [201] [202] [203] [204] [205] [206] [207] [208] [209] [210] [211] [212] [213] [214] [215] [216] [217] [218] [219] [220] [221] [222] [223] [224] [225] [226] [227] [228] [229] [230] [231] [232] [233] [234] [235] [236] [237] [238] [239] [240] [241] [242] [243] [244] [245] [246] [247] [248] [249] [250] [251] [252] [253] [254] [255] [256] [257] [258] [260] [261] [262] [263] [264] [265] [266] [267] [268] [269] [270] [271] [272] [273] [274] [275] [276] [277] [278] [279] [280] [281] [282] [283] [284] [285] [286] [287] [288] [289] [290] [291] [292] [293] [294] [295] [296] [297] [298].
3.Evidências de Catastrofismo: Revisão de marcadores geológicos de radiação intensa e impactos de asteroides sincronizados com picos de mutação observados em populações de mamíferos [122] [123] [124] [125] [126] [127] [128] [129] [130] [131] [132] [133] [134] [135] [136] [137] [138] [139] [140] [141] [142] [143] [144] [145] [146] [147] [148] [149] [150] [151] [152] [153] [154] [155] [156] [157] [158] [159] [160] [161] [300].
Resultados
O Pico Mutacional Holocênico
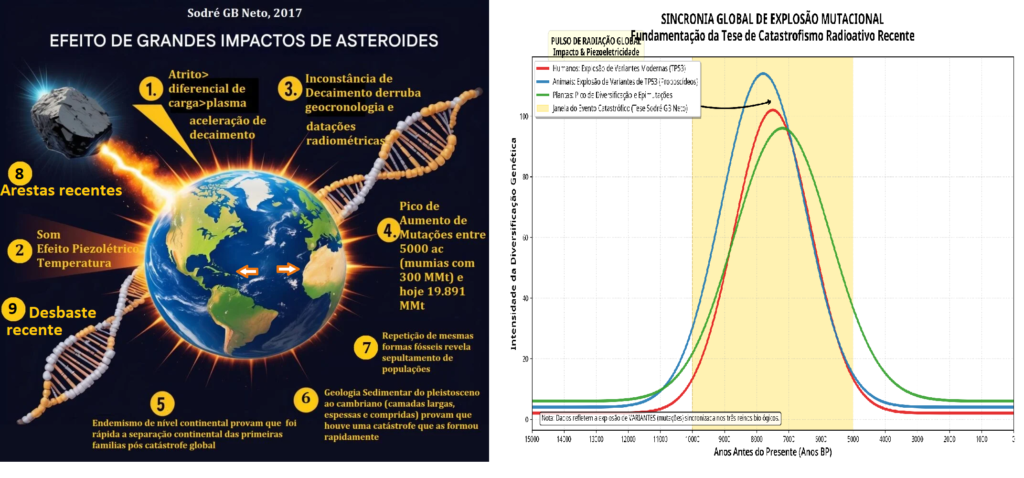
Estudos recentes sobre o DNA mitocondrial humano revelam uma discrepância alarmante [43] [45] [46] [47] [50] [52] [53] [56] [59] [60] [63] [65] [67] [68] [70] [71] [73] [74] [76] [77] [81]. Enquanto as taxas filogenéticas estimam a origem da “Eva Mitocondrial” em cerca de 150.000 a 200.000 anos, as taxas de mutação observadas diretamente em famílias (pedigree) sugerem uma cronologia de apenas 6.000 a 10.000 anos [45] [46] [63] [67] [68]. Esta divergência é explicada pela ocorrência de um evento de alta radiação no passado recente, que saturou os genomas com polimorfismos de nucleotídeo único (SNPs) [49] [51] [61] [62] [64] [66] [69] [72] [75] [78] [79] [80].
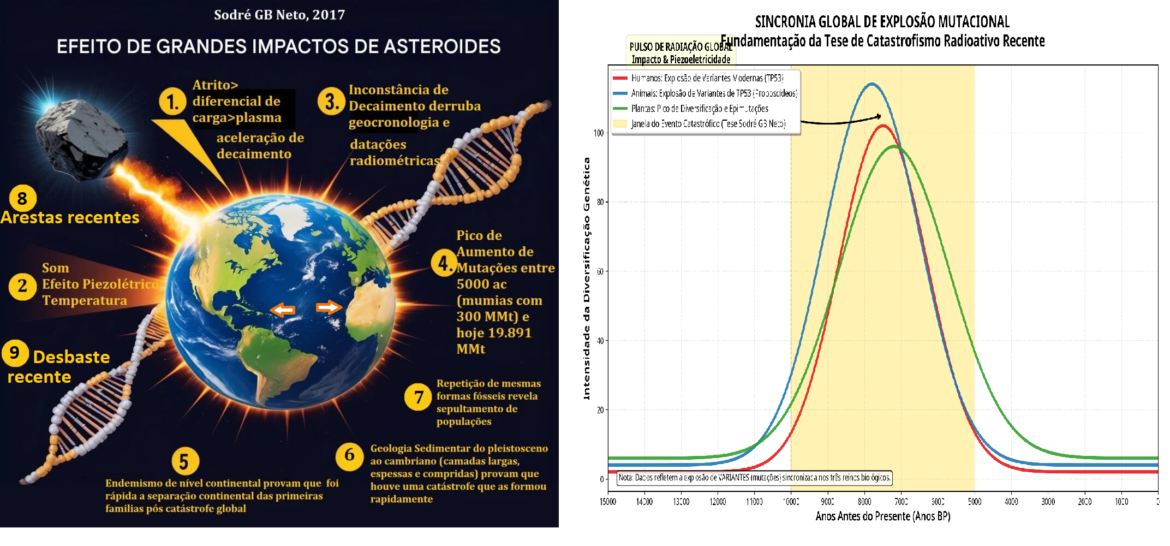
Catastrofismo Radioativo e Geocronologia
A validade da geocronologia uniformitarista é questionada pela evidência de decaimento radioativo acelerado [122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142, 143, 144, 145, 146, 147, 148, 149, 150, 151, 152, 153, 154, 155, 156, 157, 158, 159, 160, 161, 300]. Propõe-se que impactos de grandes asteroides no Holoceno geraram pressões mecânicas extremas na crosta terrestre, resultando em fenômenos de piezoeletricidade nuclear. Tais eventos liberaram fluxos massivos de nêutrons e radiação gama, alterando as proporções isotópicas usadas em datações e induzindo picos mutacionais globais [300].
Senescência Evolutiva e o Deleteriome
A acumulação de mutações não é uniforme [1, 2, 3, 4, 5]. Genes vitais para a integridade genômica, como o p53 (guardião do genoma), mostram sinais de degradação funcional [82] [83] [84] [85] [86] [87] [88] [89] [90] [91] [92] [93] [94] [95] [96] [97] [98] [99] [100] [101] [102] [103] [104] [105] [106] [107] [108] [109] [110] [111] [112] [113] [114] [115] [116] [117] [118] [119] [120] [121]. O aumento exponencial de doenças degenerativas, câncer e diabetes nas últimas décadas é um reflexo direto dessa carga genética acumulada [202] [203] [204] [205] [206] [207] [208] [209] [210] [211] [212] [213] [214] [215] [216] [217] [218] [219] [220] [221] [222] [223] [224] [225] [226] [227] [228] [229] [230] [231] [232] [233] [234] [235] [236] [237] [238] [239] [240] [241] [242] [243] [244] [245] [246] [247] [248] [249] [250] [251] [252] [253] [254] [255] [256] [257] [258] [260] [261] [262] [263] [264] [265] [266] [267] [268] [269] [270] [271] [272] [273] [274] [275] [276] [277] [278] [279] [280] [281] [282] [283] [284] [285] [286] [287] [288] [289] [290] [291] [292] [293] [294] [295] [296] [297] [298], indicando que a humanidade está atingindo um ponto de saturação mutacional [7] [13] [261].
Discussão
A integração desses conceitos revela uma narrativa científica coerente: a vida iniciou-se com genomas de alta integridade que, após um evento catastrófico recente, entraram em um processo de decaimento acelerado [122] [123] [124] [125] [126] [127] [128] [129] [130] [131] [132] [133] [134] [135] [136] [137] [138] [139] [140] [141] [142] [143] [144] [145] [146] [147] [148] [149] [150] [151] [152] [153] [154] [155] [156] [157] [158] [159] [160] [161] [300]. A “seleção natural” atua apenas como um filtro para as mutações mais grosseiramente deletérias, mas é incapaz de deter o acúmulo de milhões de variantes levemente prejudiciais que comprometem a viabilidade sistêmica [6] [7] [8] [9] [10] [11] [12] [13] [14] [15] [16] [17] [18] [19] [20] [21] [22] [23] [24] [25] [26] [27] [28] [29] [30] [31] [32] [33] [34] [35] [36] [37] [38] [39] [40] [41] [42] [48] [49] [51] [54] [55] [57] [58] [61] [62] [64] [66] [69] [72] [75] [78] [79] [80] [162] [163] [164] [165] [166] [167] [168] [169] [170] [171] [172] [173] [174] [175] [176] [177] [178] [179] [180] [181] [182] [183] [184] [185] [186] [187] [188] [189] [190] [191] [192] [193] [194] [195] [196] [197] [198] [199] [200] [201] [202] [203] [204] [205] [206] [207] [208] [209] [210] [211] [212] [213] [214] [215] [216] [217] [218] [219] [220] [221] [222] [223] [224] [225] [226] [227] [228] [229] [230] [231] [232] [233] [234] [235] [236] [237] [238] [239] [240] [241] [242] [243] [244] [245] [246] [247] [248] [249] [250] [251] [252] [253] [254] [255] [256] [257] [258] [260] [261] [262] [263] [264] [265] [266] [267] [268] [269] [270] [271] [272] [273] [274] [275] [276] [277] [278] [279] [280] [281] [282] [283] [284] [285] [286] [287] [288] [289] [290] [291] [292] [293] [294] [295] [296] [297] [298].
A invalidade da geocronologia uniformitarista é um ponto central [122] [123] [124] [125] [126] [127] [128] [129] [130] [131] [132] [133] [134] [135] [136] [137] [138] [139] [140] [141] [142] [143] [144] [145] [146] [147] [148] [149] [150] [151] [152] [153] [154] [155] [156] [157] [158] [159] [160] [161] [300]. Se as taxas de decaimento e mutação foram alteradas por eventos catastróficos, a escala de tempo da vida deve ser drasticamente reduzida [43] [45] [46] [47] [50] [52] [53] [56] [59] [60] [63] [65] [67] [68] [70] [71] [73] [74] [76] [77] [81]. Isso harmoniza a genética com o registro fóssil, onde vemos uma estase morfológica seguida de degeneração de tamanho e complexidade em muitas linhagens [122] [123] [124] [125] [126] [127] [128] [129] [130] [131] [132] [133] [134] [135] [136] [137] [138] [139] [140] [141] [142] [143] [144] [145] [146] [147] [148] [149] [150] [151] [152] [153] [154] [155] [156] [157] [158] [159] [160] [161] [300].
Conclusão
A Teoria da Degeneração Genética oferece um modelo robusto para explicar a trajetória biológica contemporânea [299]. O reconhecimento de um pico mutacional recente [45] [46] [63] [67] [68] e do acúmulo progressivo de mutações deletérias [6] [7] [8] [12] [13] [259] [261] é crucial para entender a vulnerabilidade das espécies à extinção [7] [13] [261]. A ciência deve considerar seriamente a possibilidade de que o “relógio biológico” da Terra esteja se aproximando de seu fim, exigindo uma reavaliação urgente dos paradigmas de conservação e evolução [202] [203] [204] [205] [206] [207] [208] [209] [210] [211] [212] [213] [214] [215] [216] [217] [218] [219] [220] [221] [222] [223] [224] [225] [226] [227] [228] [229] [230] [231] [232] [233] [234] [235] [236] [237] [238] [239] [240] [241] [242] [243] [244] [245] [246] [247] [248] [249] [250] [251] [252] [253] [254] [255] [256] [257] [258] [260] [261] [262] [263] [264] [265] [266] [267] [268] [269] [270] [271] [272] [273] [274] [275] [276] [277] [278] [279] [280] [281] [282] [283] [284] [285] [286] [287] [288] [289] [290] [291] [292] [293] [294] [295] [296] [297] [298].
Referências
14.Eco-evolutionary extinction and recolonization dynamics reduce genetic load and increase time to extinction in highly inbred populations.. PMID: 36117269, PMC: PMC9828521. Link: https://pubmed.ncbi.nlm.nih.gov/36117269/
33.Genetic variability and founder effect in the pitcher plant Sarracenia purpurea (Sarraceniaceae ) in populations introduced into Switzerland: from inbreeding to invasion.. PMID: 15546932, PMC: PMC4246826. Link: https://pubmed.ncbi.nlm.nih.gov/15546932/
43.Genetic Diversity Analysis of the Chinese Daur Ethnic Group in Heilongjiang Province by Complete Mitochondrial Genome Sequencing.. PMID: 35801081, PMC: PMC9253502. Link: https://pubmed.ncbi.nlm.nih.gov/35801081/
44.Containing the spread of COVID-19 virus facing to its high mutation rate: approach to intervention using a nonspecific way of blocking its entry into the cells.. PMID: 35532338. Link: https://pubmed.ncbi.nlm.nih.gov/35532338/
46.Human molecular evolutionary rate, time dependency and transient polymorphism effects viewed through ancient and modern mitochondrial DNA genomes.. PMID: 33658608, PMC: PMC7930196. Link: https://pubmed.ncbi.nlm.nih.gov/33658608/
49.Mutational signatures of redox stress in yeast single-strand DNA and of aging in human mitochondrial DNA share a common feature.. PMID: 31067233, PMC: PMC6527239. Link: https://pubmed.ncbi.nlm.nih.gov/31067233/
54.Reduced Mitochondrial Membrane Potential Is a Late Adaptation of Trypanosoma brucei brucei to Isometamidium Preceded by Mutations in the γ Subunit of the F1Fo-ATPase.. PMID: 27518185, PMC: PMC4982688. Link: https://pubmed.ncbi.nlm.nih.gov/27518185/
59.Examining phylogenetic relationships among gibbon genera using whole genome sequence data using an approximate bayesian computation approach.. PMID: 25769979, PMC: PMC4423371. Link: https://pubmed.ncbi.nlm.nih.gov/25769979/
64.Bioenergetics in human evolution and disease: implications for the origins of biological complexity and the missing genetic variation of common diseases.. PMID: 23754818, PMC: PMC3685467. Link: https://pubmed.ncbi.nlm.nih.gov/23754818/
73.Phylogeography, genetic structure and population divergence time of cheetahs in Africa and Asia: evidence for long-term geographic isolates.. PMID: 21214655, PMC: PMC3531615. Link: https://pubmed.ncbi.nlm.nih.gov/21214655/
81.Investigation of heteroplasmy in the human mitochondrial DNA control region: a synthesis of observations from more than 5000 global population samples.. PMID: 19407924. Link: https://pubmed.ncbi.nlm.nih.gov/19407924/
82.Real-world disease characteristics and frontline treatments used in chronic lymphocytic leukaemia in Bulgaria: An observational cohort study (DESCRIBE ).. PMID: 41698655. Link: https://pubmed.ncbi.nlm.nih.gov/41698655/
85.Histological and Genetic Markers of Cellular Senescence in Keratinocyte Cancers and Actinic Keratosis: A Systematic Review.. PMID: 41683940, PMC: PMC12898786. Link: https://pubmed.ncbi.nlm.nih.gov/41683940/
86.Multi-gene co-mutations of BRAF with TERT, PIK3CA, or TP53 are powerful predictors of central lymph node metastasis in papillary thyroid carcinoma.. PMID: 41675640, PMC: PMC12886009. Link: https://pubmed.ncbi.nlm.nih.gov/41675640/
93.Alignment of Molecular Classification Between Diagnosis and Recurrence in Endometrial Cancer: Lessons from a Single-Institution Experience to Inform Future Pathways.. PMID: 41595167, PMC: PMC12839114. Link: https://pubmed.ncbi.nlm.nih.gov/41595167/
95.Clinicopathological characteristics and therapeutic outcomes in patients with non-small cell lung cancer harboring SMARCA4 mutations.. PMID: 41593906, PMC: PMC12854217. Link: https://pubmed.ncbi.nlm.nih.gov/41593906/
100.Mismatch Repair Deficiency Profiling and Its Impact on Management and Prognosis in Endometrial Cancer Patients: A Comprehensive Update.. PMID: 41552063, PMC: PMC12805798. Link: https://pubmed.ncbi.nlm.nih.gov/41552063/
104.Clinical impact of TP53 classifications in previously treated advanced driver-negative non-small cell lung cancer: A biomarker analysis of the OAK and POPLAR randomized clinical trials.. PMID: 41500083. Link: https://pubmed.ncbi.nlm.nih.gov/41500083/
108.Molecular profile-based adjuvant treatment for women with high-intermediate risk endometrial cancer (PORTEC-4a ): results of a randomised, open-label, phase 3, multicentre, non-inferiority trial.. PMID: 41449145. Link: https://pubmed.ncbi.nlm.nih.gov/41449145/
111.Association between molecular classification and overall survival in patients with metastatic endometrial carcinoma: ancillary results of the UTOLA phase II GINECO trial.. PMID: 41418627. Link: https://pubmed.ncbi.nlm.nih.gov/41418627/
112.Characterization of chromosome 5 aberrations in TP53 mutated myeloid neoplasms with ≥5% blasts: An International TP53 Investigators Network (iTiN ) study.. PMID: 41417597, PMC: PMC12716622. Link: https://pubmed.ncbi.nlm.nih.gov/41417597/
115.Next-Generation Sequencing Reveals a Diagnostic and Prognostic Role of the TP53 R273C Mutation in Lower-Grade, IDH-Mutant Astrocytomas.. PMID: 41373636, PMC: PMC12692303. Link: https://pubmed.ncbi.nlm.nih.gov/41373636/
119.Whole-exome sequencing for identification of specific gene mutations in an Indian cohort of triple-negative breast cancer patients.. PMID: 41324770, PMC: PMC12775201. Link: https://pubmed.ncbi.nlm.nih.gov/41324770/
121.HER2 immunoreactivity in advanced non-p53abn endometrial carcinoma: Association with clinical features, prognosis, and molecular characteristics.. PMID: 41314003. Link: https://pubmed.ncbi.nlm.nih.gov/41314003/
125.Quaternary climatic oscillations shaped the demographic history and triggered intraspecific divergence of Rhododendron shanii, a mid-montane endemic in eastern Asia.. PMID: 41602536, PMC: PMC12833463. Link: https://pubmed.ncbi.nlm.nih.gov/41602536/
126.Elucidating Continental-Wide Phylogeographic and Adaptive Processes Shaping the Genome-Wide Diversity of North America’s Most Widely Distributed Tree.. PMID: 41324157, PMC: PMC12717988. Link: https://pubmed.ncbi.nlm.nih.gov/41324157/
133.Population genomics of flat-tailed horned lizards (Phrynosoma mcallii ) informs conservation and management across a fragmented Colorado Desert landscape.. PMID: 38445567. Link: https://pubmed.ncbi.nlm.nih.gov/38445567/
134.Exploring the genetic diversity and population structure of Ailanthus altissima using chloroplast and nuclear microsatellite DNA markers across its native range.. PMID: 38078105, PMC: PMC10702488. Link: https://pubmed.ncbi.nlm.nih.gov/38078105/
143.Inferring historical survivals of climate relicts: the effects of climate changes, geography, and population-specific factors on herbaceous hydrangeas.. PMID: 33510468, PMC: PMC8115046. Link: https://pubmed.ncbi.nlm.nih.gov/33510468/
144.Population genetics and evolutionary history of the endangered Eld’s deer (Rucervus eldii ) with implications for planning species recovery.. PMID: 33510319, PMC: PMC7844053. Link: https://pubmed.ncbi.nlm.nih.gov/33510319/
147.Insights into the neutral and adaptive processes shaping the spatial distribution of genomic variation in the economically important Moroccan locust (Dociostaurus maroccanus ).. PMID: 32489626, PMC: PMC7244894. Link: https://pubmed.ncbi.nlm.nih.gov/32489626/
153.Demographic inference in barn swallows using whole-genome data shows signal for bottleneck and subspecies differentiation during the Holocene.. PMID: 30176075. Link: https://pubmed.ncbi.nlm.nih.gov/30176075/
155.History of the fragmentation of the African rain forest in the Dahomey Gap: insight from the demographic history of Terminalia superba.. PMID: 29279603, PMC: PMC5943585. Link: https://pubmed.ncbi.nlm.nih.gov/29279603/
156.Did Late Pleistocene climate change result in parallel genetic structure and demographic bottlenecks in sympatric Central African crocodiles, Mecistops and Osteolaemus?. PMID: 29024142. Link: https://pubmed.ncbi.nlm.nih.gov/29024142/
157.Whole Mitogenomes Reveal the History of Swamp Buffalo: Initially Shaped by Glacial Periods and Eventually Modelled by Domestication.. PMID: 28680070, PMC: PMC5498497. Link: https://pubmed.ncbi.nlm.nih.gov/28680070/
169.Multiple Genetic Impacts of Immigration Interact to Shape Local Population Persistence versus Extinction: Evolutionary Rescue, Inbreeding Vortex, and Migrational Meltdown.. PMID: 41349120. Link: https://pubmed.ncbi.nlm.nih.gov/41349120/
171.Attenuation of HIV severity by slightly deleterious mutations can explain the long-term trajectory of virulence evolution.. PMID: 41325373, PMC: PMC12677797. Link: https://pubmed.ncbi.nlm.nih.gov/41325373/
173.Composition of the Gut Microbiome and Its Response to Rice Stripe Virus Infection in Laodelphax striatellus (Hemiptera: Delphacidae ).. PMID: 41302881, PMC: PMC12653645. Link: https://pubmed.ncbi.nlm.nih.gov/41302881/
179.Genomic and population genomic analyses reveal contrasting diversity and demographic histories in a critically endangered and a widespread Oreocharis species.. PMID: 41054605, PMC: PMC12496536. Link: https://pubmed.ncbi.nlm.nih.gov/41054605/
180.Genetic factors may load the gun, but environmental factors pull the trigger: MedDiet and DII in rheumatoid arthritis.. PMID: 41041138, PMC: PMC12483893. Link: https://pubmed.ncbi.nlm.nih.gov/41041138/
182.Genome sequencing and population genomics provide insights into the demographic history, genetic load, and local adaptation of an endangered Tertiary relict.. PMID: 40827714. Link: https://pubmed.ncbi.nlm.nih.gov/40827714/
183.SARS-CoV-2 genomic diversity and within-host evolution in individuals with persistent infection in the UK: an observational, longitudinal, population-based surveillance study.. PMID: 40752506. Link: https://pubmed.ncbi.nlm.nih.gov/40752506/
184.Population Genomic Analysis Provides Insights Into the Evolution and Conservation of Two Critically Endangered Musk Deer Species.. PMID: 40747002, PMC: PMC12311986. Link: https://pubmed.ncbi.nlm.nih.gov/40747002/
187.Effective Purging and Conservation of Heterozygous Regions During Independent Evolution to High-Level Inbreeding in Wild Paradise Fishes.. PMID: 40686397. Link: https://pubmed.ncbi.nlm.nih.gov/40686397/
189.Multi-ancestry genome-wide meta-analysis of 56,241 individuals identifies known and novel cross-population and ancestry-specific associations as novel risk loci for Alzheimer’s disease.. PMID: 40676597, PMC: PMC12273372. Link: https://pubmed.ncbi.nlm.nih.gov/40676597/
198.Natural dispersal is better than translocation for reducing risks of inbreeding depression in eastern black rhinoceros (Diceros bicornis michaeli ).. PMID: 40460127, PMC: PMC12167989. Link: https://pubmed.ncbi.nlm.nih.gov/40460127/
201.Conservation genomics of a threatened subtropical Rhododendron species highlights the distinct conservation actions required in marginal and admixed populations.. PMID: 40287966, PMC: PMC12034323. Link: https://pubmed.ncbi.nlm.nih.gov/40287966/
208.Under the Shadow: Old-biased Genes Are Subject to Weak Purifying Selection at Both the Tissue- and Cell Type-Specific Levels.. PMID: 41092135, PMC: PMC12569596. Link: https://pubmed.ncbi.nlm.nih.gov/41092135/
210.Alterations in MRI-visible perivascular spaces precede dementia diagnosis by 18 years in autosomal dominant Alzheimer’s disease.. PMID: 40851076, PMC: PMC12375433. Link: https://pubmed.ncbi.nlm.nih.gov/40851076/
224.The light-harvesting chlorophyll a/b-binding proteins of photosystem II family members are responsible for temperature sensitivity and leaf color phenotype in albino tea plant.. PMID: 38151116, PMC: PMC11674787. Link: https://pubmed.ncbi.nlm.nih.gov/38151116/
229.Expanding evolutionary theories of ageing to better account for symbioses and interactions throughout the Web of Life.. PMID: 37321383, PMC: PMC10771319. Link: https://pubmed.ncbi.nlm.nih.gov/37321383/
232.Mitochondrial effects on fertility and longevity in Tigriopus californicus contradict predictions of the mother’s curse hypothesis.. PMID: 36382523, PMC: PMC9667352. Link: https://pubmed.ncbi.nlm.nih.gov/36382523/
233.On the illusion of auxotrophy: met15Δ yeast cells can grow on inorganic sulfur, thanks to the previously uncharacterized homocysteine synthase Yll058w.. PMID: 36379252, PMC: PMC9763685. Link: https://pubmed.ncbi.nlm.nih.gov/36379252/
235.Multigenerational downregulation of insulin/IGF-1 signaling in adulthood improves lineage survival, reproduction, and fitness in Caenorhabditis elegans supporting the developmental theory of ageing.. PMID: 36199198, PMC: PMC10092551. Link: https://pubmed.ncbi.nlm.nih.gov/36199198/
236.A quantification method of somatic mutations in normal tissues and their accumulation in pediatric patients with chemotherapy.. PMID: 35895679, PMC: PMC9351471. Link: https://pubmed.ncbi.nlm.nih.gov/35895679/
242.A fixed mutation in the respiratory complex I impairs mitochondrial bioenergetics in the endangered Apennine brown bear.. PMID: 41026818, PMC: PMC12519208. Link: https://pubmed.ncbi.nlm.nih.gov/41026818/
244.Genomic evidence for demographic fluctuations, genetic burdens and adaptive divergence in fourfinger threadfin Eleutheronema rhadinum.. PMID: 40027332, PMC: PMC11871173. Link: https://pubmed.ncbi.nlm.nih.gov/40027332/
248.How should we measure population-level inbreeding depression? Impacts of standing genetic associations between selfing rate and deleterious mutations.. PMID: 39045597, PMC: PMC11263115. Link: https://pubmed.ncbi.nlm.nih.gov/39045597/
253.Genome sequences and population genomics provide insights into the demographic history, inbreeding, and mutation load of two ‘living fossil’ tree species of Dipteronia.. PMID: 37797086. Link: https://pubmed.ncbi.nlm.nih.gov/37797086/
260.An evolutionary perspective on genetic load in small, isolated populations as informed by whole genome resequencing and forward-time simulations.. PMID: 36626799. Link: https://pubmed.ncbi.nlm.nih.gov/36626799/
264.Mitonuclear Mismatch is Associated With Increased Male Frequency, Outcrossing, and Male Sperm Size in Experimentally-Evolved C. elegans.. PMID: 35360860, PMC: PMC8961728. Link: https://pubmed.ncbi.nlm.nih.gov/35360860/
273.The evolution of metapopulation dynamics and the number of stem cells in intestinal crypts and other tissue structures in multicellular bodies.. PMID: 32821281, PMC: PMC7428809. Link: https://pubmed.ncbi.nlm.nih.gov/32821281/
281.A chromosome-scale genome and transcriptomic analysis of the endangered tropical tree Vatica mangachapoi (Dipterocarpaceae ).. PMID: 35171284, PMC: PMC8882376. Link: https://pubmed.ncbi.nlm.nih.gov/35171284/
288.Two-dimensional IR spectroscopy of the anti-HIV agent KP1212 reveals protonated and neutral tautomers that influence pH-dependent mutagenicity.. PMID: 25733867, PMC: PMC4371980. Link: https://pubmed.ncbi.nlm.nih.gov/25733867/
289.Tautomerism provides a molecular explanation for the mutagenic properties of the anti-HIV nucleoside 5-aza-5,6-dihydro-2′-deoxycytidine.. PMID: 25071207, PMC: PMC4136561. Link: https://pubmed.ncbi.nlm.nih.gov/25071207/
299.Genetic Entropy and the Mystery of the Genome.. ISBN: 978-0981631608. (Sanford, 2008 ).