January 2026
DOI:10.13140/RG.2.2.15799.38563 . Sodré Gonçalves de Brito Neto
Resumo: A acumulação acelerada de mutações no genoma humano entre 5.000 e 10.000 anos atrás tem sido erroneamente atribuída por Gerald Crabtree e por boa parte do consenso científico, a mudanças no estilo de vida de caçador-coletor sob forte pressão seletiva, para agricultor, o que protegeria os mais fracos e doentes para que acumulassem mais defeitos genéticos nas descendencias. Este artigo propõe uma mudança deste paradigma, porque o pico de mutações observado em humanos modernos, também é observado em diversos animais; onde temos entre vários exemplos, o trecho genético TP53 de proboscídeos fósseis quando comparados a elefantes modernos que explodem variações mutadas, assim como ocorre no homem moderno comparado a alguns neandertais , requerendo assim uma causa não cultural invalidando a hipótese da transição de caçador-coletor para agricultor, porque elefantes e dezenas de outros animais não se tornaram agrocultores; esta causa afetaria humanos e animais igualmente e portanto seria global e catastrófica, sendo portanto um acontecimento acelerador de decaimento radioativo. Deduzimos que este pico mutacional recente foi resultado direto de um evento catastrófico acelerador de decaimento radioativo. Através da aplicação da teoria da piezoeletricidade nuclear de Cardone e Carpinteri , argumentamos que se fraturas de rochas e terremotos geram acekleração de decaimento e liberação de neutrons, quanto mais impactos de grandes asteroides que fabricariam milhares de terremotos, vibrando todfa terra; geraram pressões na escala de Gigapascals, induzindo a liberação de nêutrons em larga escala e fono-fissão. Também a prevalência de polimorfismos de nucleotídeo único (SNPs) no gene TP53 humano e DNAmt, pode ser interpretada como uma assinatura genômica de eventos mutagênicos intensos e generalizados na história entrópica humana. Contrariamente à hipótese do consenso acadêmico citada por Crabtree, que postula um ‘intelecto frágil’ resultante de uma diminuição da pressão seletiva cultural, propomos que a acumulação de tais variações no TP53, e em outros genes críticos, pode ser mais plausivelmente atribuída principalmente a uma causa catastrófica global de natureza radioativa. Tal evento teria imposto uma pressão mutagênica sem precedentes, explicando entre muitos aspectos o alto contraste genético e de tamanho médio, entre ancestrais fosseis e sobreviventes na biodiversidade atual ; levando a uma rápida diversificação genômica e à fixação de SNPs que, embora pudessem conferir alguma adaptabilidade em um ambiente pós-catastrófico, também poderiam ter contribuído para a vulnerabilidade intrínseca dos humanos modernos a doenças como o câncer e queda drástica de longevidade já que TP53 como reparador celular está estreitamente relacionado a longevidade . A alta frequência de SNPs no TP53, portanto, não reflete uma fragilidade intelectual culturalmente induzida, mas sim uma cicatriz molecular de um passado geológico e ambiental tumultuado, moldando a biologia humana de maneiras profundas e duradouras.
Introdução
Materiais e Métodos
Resultados e Discussão
Piezoeletricidade Nuclear e a Invalidação da Geocronologia
Efeitos verificados na queda de grandes bólidos como “Espallação”, piezoeletricidade nuclear (Carpinteri, h= 95[7]), fono-fissão [17], plasmas de altíssimas amperagens e diferenciais de carga promovem decaimento acelerado, alterando a constância de decaimento, podendo “envelhecer” rochas em milissegundos falseando a datação radiométrica uniformitarianistas.
A teoria da piezoeletricidade nuclear demonstra que pressões extremas e ondas de choque mecânicas podem induzir reações nucleares até mesmo sem a necessidade de altas temperaturas (fissão piezonuclear) . Cardone et al. demonstraram a aceleração do decaimento do Tório sob cavitação acústica, um fenómeno que sugere que a taxa de decaimento não é uma constante eterna, mas dependente do ambiente físico-químico e mecânico.
O Pico Mutacional como Subproduto de Impactos Nucleares
Geologia de Catástrofe e Estratificação Spontânea
Tabela de Evolução do Gene TP53 em Mamíferos: Do Canônico ao Variável
|
#
|
Ancestral (Fóssil/Reconstruído)
|
Descendente Moderno
|
Estado Ancestral (NM_000546)
|
Variações no Descendente Moderno
|
Referência (DOI/PMID)
|
|---|---|---|---|---|---|
|
1
|
Neandertal
|
Homem Moderno
|
Canônico
|
~1000 variações (ex: P72R, R248W)
|
|
|
2
|
Mamute Lanoso
|
Elefante Africano
|
Canônico
|
Expansão para 20 cópias (1 gene + 19 retrogenes)
|
|
|
3
|
Basilosauridae
|
Baleia-franca
|
Canônico
|
Substituição Leu na região rica em prolinas
|
|
|
4
|
Ancestral Quiróptero
|
Morcego-de-Brandt
|
Canônico
|
Inserção de 7 aa na região de ligação ao DNA
|
|
|
5
|
Ancestral Roedor
|
Rato-toupeira-pelado
|
Canônico
|
Estabilização extrema e acúmulo nuclear
|
|
|
6
|
Ancestral Cetáceo
|
Baleia-azul
|
Canônico
|
Seleção positiva em vias de supressão tumoral
|
|
|
7
|
Ancestral Fiseterídeo
|
Cachalote
|
Canônico
|
Variações em genes da via p53 (Peto’s Paradox)
|
|
|
8
|
Ancestral Delfinídeo
|
Golfinho-nariz-de-garrafa
|
Canônico
|
Seleção positiva em resíduos conservados
|
|
|
9
|
Ancestral Sirênio
|
Peixe-boi
|
Canônico
|
Expansão de cópias de TP53
|
|
|
10
|
Ancestral Spalacídeo
|
Rato-toupeira-cego
|
Canônico
|
Substituição Arg174Lys (afinidade ao DNA)
|
|
|
11
|
Ancestral Hominídeo
|
Chimpanzé
|
Canônico
|
Diferenças na regulação transcricional
|
|
|
12
|
Ancestral Hominídeo
|
Gorila
|
Canônico
|
Variações na região promotora
|
|
|
13
|
Urso Ancestral
|
Urso Polar
|
Canônico
|
Seleção positiva em genes de reparo de DNA
|
|
|
14
|
Ancestral Pinípede
|
Foca-de-baikal
|
Canônico
|
Adaptações para hipóxia na via p53
|
|
|
15
|
Ancestral Quiróptero
|
Morcego-pequeno-marrom
|
Canônico
|
Inserções na região de ligação ao DNA
|
|
|
16
|
Ancestral Esquilo
|
Esquilo-terrestre
|
Canônico
|
Variações ligadas à hibernação
|
|
|
17
|
Ancestral Camelídeo
|
Camelo
|
Canônico
|
Seleção positiva em resposta ao estresse
|
|
|
18
|
Ancestral Girafídeo
|
Girafa
|
Canônico
|
Adaptações no ciclo celular (pressão alta)
|
|
|
19
|
Ancestral Rinoceronte
|
Rinoceronte-branco
|
Canônico
|
Variações em supressores de tumor
|
|
|
20
|
Ancestral Xenarthra
|
Tatu-galinha
|
Canônico
|
Duplicação massiva de genes supressores
|
|
|
21
|
Ancestral Pilosa
|
Preguiça-de-dois-dedos
|
Canônico
|
Proliferação celular lenta
|
|
|
22
|
Ancestral Pilosa
|
Tamanduá-bandeira
|
Canônico
|
Duplicação de genes da via p53
|
|
|
23
|
Ancestral Monotremado
|
Ornitorrinco
|
Canônico
|
Traços ancestrais de répteis
|
|
|
24
|
Ancestral Monotremado
|
Equidna
|
Canônico
|
Variações genômicas únicas
|
|
|
25
|
Ancestral Marsupial
|
Diabo-da-tasmânia
|
Canônico
|
Seleção positiva (tumor facial)
|
|
|
26
|
Ancestral Marsupial
|
Canguru-vermelho
|
Canônico
|
Variações em genes de reparo de DNA
|
|
|
27
|
Ancestral Marsupial
|
Gambá-de-orelha-preta
|
Canônico
|
Conservação com variações específicas
|
|
|
28
|
Ancestral Sirênio
|
Peixe-boi-da-amazônia
|
Canônico
|
Expansão de cópias de TP53
|
|
|
29
|
Ancestral Sirênio
|
Dugongo
|
Canônico
|
Variações em genes supressores
|
|
|
30
|
Ancestral Proboscídeo
|
Elefante Asiático
|
Canônico
|
Expansão de retrogenes TP53
|
|
|
31
|
Ancestral Bovídeo
|
Vaca
|
Canônico
|
Retroposon antigo no promotor de TP53
|
|
|
32
|
Ancestral Canídeo
|
Cão
|
Canônico
|
Variações em hotspots de mutação de p53
|
Envelhecimetno e P53
O envelhecimento humano é um processo complexo caracterizado pelo declínio progressivo das funções fisiológicas e pela perda da homeostase molecular. A proteína p53, conhecida como o “guardião do genoma”, desempenha um papel central na regulação do ciclo celular, reparo do DNA e apoptose [1]. No entanto, ao atingir a fase idosa, particularmente após os 60 anos, observa-se uma diminuição significativa na funcionalidade da p53 em homens [2]. Este fenômeno coincide com a andropausa e o declínio de diversas enzimas e proteínas essenciais [3]. Este artigo revisa os mecanismos moleculares subjacentes à perda de função da p53, a influência do declínio hormonal e as consequências para a estabilidade genômica e a longevidade [4] [36] [37] [38] [39] [40] [41] [42] [48] [49] [50] [51] [52] [53] [54] [55].
Introdução
A p53 é um fator de transcrição ativado em resposta a estresses celulares, como dano ao DNA, hipóxia e estresse oncogênico [5]. Sua função primordial é integrar esses sinais para determinar o destino celular: reparo e sobrevivência, senescência estável ou morte celular programada (apoptose) [6] [56] [57] [58] [59] [60] [61] [62] [63] [64] [65] [66] [67] [68] [69] [70] [71] [72] [73] [74] [75] [76] [77] [78] [79] [80] [81] [82] [83] [84] [85] [86] [87] [88] [89] [90] [91] [92] [93] [94] [95] [96] [97] [98] [99] [100] [101] [102] [103]. Em indivíduos jovens, a p53 induz a parada reversível do ciclo celular em níveis baixos de ativação, permitindo o reparo do DNA [7].
Com o avanço da idade, a eficácia dessa resposta protetora diminui, o que é um fator contribuinte para o aumento da incidência de câncer e doenças relacionadas ao envelhecimento em populações idosas [8]. Em homens acima de 60 anos, o declínio da testosterona e a alteração no perfil enzimático contribuem para um ambiente celular que favorece a inativação ou a instabilidade da p53 [9] [10].
Mecanismos de Perda de Funcionalidade da p53
A perda de função da p53 no envelhecimento é multifatorial e transcende a simples aquisição de mutações somáticas no gene TP53 [11]. Embora mutações no TP53 sejam a alteração genética mais comum em cânceres humanos, a perda de funcionalidade no envelhecimento saudável está frequentemente ligada a mecanismos pós-traducionais e regulatórios [12] [54] [55].
1.Instabilidade e Degradação Proteica: Estudos indicam que a estabilização da proteína p53 após o estresse é reduzida em tecidos envelhecidos [13]. O desequilíbrio na proteostase, característico do envelhecimento, leva à agregação de proteínas e à perda de função de organelas celulares, como o retículo endoplasmático e as mitocôndrias [14]. A interação com chaperonas moleculares, como as proteínas de choque térmico (HSPs), que podem estabilizar a p53 mutante, também se torna desregulada [15] [56].
2.Alteração na Dinâmica de Sinalização: A decisão celular mediada pela p53 é regida pela amplitude e duração de sua ativação [16] [57]. A dinâmica da p53 — oscilatória versus respostas sustentadas — reflete como as células integram a intensidade e a persistência do dano ao DNA [17]. Com o envelhecimento, essa dinâmica é alterada, resultando em uma falha na indução de programas de senescência ou apoptose quando necessários, favorecendo a sobrevivência de células danificadas [18] [58] [59] [60] [61] [62] [63] [64] [65] [66] [67] [68] [69] [70] [71] [72] [73] [74] [75] [76] [77] [78] [79] [80] [81] [82] [83] [84] [85] [86] [87] [88] [89] [90] [91] [92] [93] [94] [95] [96] [97] [98] [99] [100] [101] [102] [103].
3.Isoformas de p53: A expressão diferencial das isoformas de p53 (como p53β, Δ133p53α e Δ40p53) funciona como um “reostato” que ajusta a sensibilidade da célula à senescência [19]. O balanço entre essas isoformas é alterado com a idade, contribuindo para a heterogeneidade da resposta celular ao estresse [20] [37].
O Papel da Andropausa e do Declínio Enzimático
A partir dos 60 anos, o homem entra em uma fase de declínio hormonal progressivo, conhecida como andropausa ou hipogonadismo tardio [21]. A redução dos níveis de testosterona tem sido associada à diminuição da expressão de genes regulados pela p53 e ao aumento do estresse oxidativo [22] [23]. A testosterona, por exemplo, pode modular a fosforilação da p53 em resposta ao estresse oxidativo [24] [60] [61] [62] [63] [64] [65].
Simultaneamente, enzimas metabólicas e mitocondriais apresentam queda em sua atividade [25]. A p53 é um regulador mestre do metabolismo, controlando enzimas como a Glutaminase 2 (GLS2), essencial para a produção de energia e defesa antioxidante [26] [43] [44] [45] [46] [47]. O declínio na atividade de enzimas mitocondriais, como as da cadeia respiratória, é um biomarcador do envelhecimento [27], e a p53 regula a expressão de proteínas mitocondriais [28]. Esse declínio enzimático cria um ciclo de feedback positivo onde o aumento do dano oxidativo inativa ainda mais a p53, comprometendo a capacidade de reparo e defesa antioxidante [29] [66] [67] [68] [69] [70] [71] [72] [73] [74] [75] [76] [77] [78] [79] [80] [81] [82] [83] [84] [85] [86] [87] [88] [89] [90] [91] [92] [93] [94] [95] [96] [97] [98] [99] [100] [101] [102] [103].
Consequências Sistêmicas
A perda de funcionalidade da p53 em homens idosos resulta em um acúmulo de células senescentes que secretam fatores pró-inflamatórios (SASP), contribuindo para a inflamação crônica de baixo grau, ou “inflammaging” [30] [31] [42] [77] [78] [79] [80] [81] [82] [83] [84] [85] [86] [87] [88] [89] [90] [91] [92] [93] [94] [95] [96] [97] [98] [99] [100] [101] [102] [103]. Esse estado inflamatório crônico predispõe o organismo a doenças degenerativas, como neurodegeneração, doenças cardiovasculares e o câncer [32] [33]. A falha da p53 em induzir a apoptose ou senescência de células danificadas permite a sobrevivência de células com instabilidade genômica, acelerando a tumorigênese [34].
A perda de funcionalidade da p53 após os 60 anos em homens é um evento molecular crucial que se interliga com o declínio hormonal da andropausa e a disfunção enzimática metabólica. Essa tríade de fatores compromete a estabilidade genômica e a homeostase tecidual, contribuindo significativamente para o fenótipo do envelhecimento e o aumento da suscetibilidade a doenças. A compreensão detalhada desses mecanismos é fundamental para o desenvolvimento de senolíticos e senomórficos que visam restaurar a função da p53 ou eliminar células senescentes, promovendo um envelhecimento saudável [35] [40] [42] [77] [78] [79] [80] [81] [82] [83] [84] [85] [86] [87] [88] [89] [90] [91] [92] [93] [94] [95] [96] [97] [98] [99] [100] [101] [102] [103].
Referências A
1. Kring, D. A. (2007). The Chicxulub impact event and its environmental consequences. Chemie der Erde – Geochemistry, 67(1), 1–36. DOI: [https://doi.org/10.1016/j.chemer.2007.04.002](https://doi.org/10.1016/j.chemer.2007.04.002).
2. Morgan, J. V., et al. (2022). The Chicxulub impact and its environmental consequences. Nature Reviews Earth & Environment, 3(4), 232–246. DOI: [https://doi.org/10.1038/s43017-022-00283-y](https://doi.org/10.1038/s43017-022-00283-y).
3. Collins, G. S., et al. (2008). A numerical study of the formation of the Vredefort impact structure. Meteoritics & Planetary Science, 43(12), 1955–1966. DOI: [https://doi.org/10.1111/j.1945-5100.2008.tb00644.x](https://doi.org/10.1111/j.1945-5100.2008.tb00644.x).
4. Navarro, K. F., et al. (2020). Emission spectra of a simulated Chicxulub impact-vapor plume. Icarus, 345, 113735. DOI: [https://doi.org/10.1016/j.icarus.2020.113735](https://doi.org/10.1016/j.icarus.2020.113735).
5. Kletetschka, G., et al. (2021). Plasma shielding removes prior magnetization record from impact melt. Scientific Reports, 11(1), 1–10. DOI: [https://doi.org/10.1038/s41598-021-01451-8](https://doi.org/10.1038/s41598-021-01451-8).
6. Leckenby, G., et al. (2024). High-temperature 205Tl decay clarifies 205Pb dating in early solar system. Nature Communications, 15(1), 1–11. PMC: PMC11560843. DOI: [https://doi.org/10.1038/s41467-024-54179-w](https://doi.org/10.1038/s41467-024-54179-w).
7. Mishra, B., et al. (2023). Plasma $\beta$-Decay Rates in the Framework of PANDORA Project. EPJ Web of Conferences, 288, 02001. DOI: [https://doi.org/10.1051/epjconf/202328802001](https://doi.org/10.1051/epjconf/202328802001).
8. Emery, G. T. (1972). Perturbation of nuclear decay rates. Annual Review of Nuclear Science, 22(1), 165–202. DOI: [https://doi.org/10.1146/annurev.ns.22.120172.001121](https://doi.org/10.1146/annurev.ns.22.120172.001121).
9. Timashev, S. F. (2015). Radioactive decay as a forced nuclear chemical process: Phenomenology. Russian Journal of Physical Chemistry A, 89(11), 1903–1910.
10. Fischbach, E., et al. (2009). Time-dependent nuclear decay parameters: new evidence for new forces?. Space Science Reviews, 145(3-4), 285–305. DOI: [https://doi.org/10.1007/s11214-009-9518-5](https://doi.org/10.1007/s11214-009-9518-5).
11. Pálffy, A., et al. (2020). Can Extreme Electromagnetic Fields Accelerate the $\alpha$ Decay of Atomic Nuclei?. Physical Review Letters, 124(21), 212505. DOI: [https://doi.org/10.1103/PhysRevLett.124.212505](https://doi.org/10.1103/PhysRevLett.124.212505).
12. Carpinteri, A., & Manuello, A. (2011). Geomechanical and Geochemical Evidence of Piezonuclear Fission Reactions in the Earth’s Crust. Strain, 47(s2), 267–281. DOI: [https://doi.org/10.1111/j.1475-1305.2010.00766.x](https://doi.org/10.1111/j.1475-1305.2010.00766.x).
13. Carpinteri, A., Lacidogna, G., & Manuello, A. (2012). Piezonuclear Fission Reactions in Rocks: Evidences from Microchemical Analysis, Neutron Emission, and Geological Transformation. Rock Mechanics and Rock Engineering, 45(4), 621–633. DOI: [https://doi.org/10.1007/s00603-011-0217-7](https://doi.org/10.1007/s00603-011-0217-7).
14. Allen, N. H., et al. (2022). A Revision of the Formation Conditions of the Vredefort Crater. Journal of Geophysical Research: Planets, 127(5), e2022JE007186. DOI: [https://doi.org/10.1029/2022JE007186](https://doi.org/10.1029/2022JE007186).
15. Taleyarkhan, R. P., et al. (2002). Evidence for nuclear emissions during acoustic cavitation. Science, 295(5561), 1868–1873. DOI: [https://doi.org/10.1126/science.1067589](https://doi.org/10.1126/science.1067589). PMID: 11884748.
- Davis, C.L. (2016). Geocronologia Microestrutural do Zircão Através da Elevação Central da Estrutura de Impacto de Vredefort. Electronic Thesis and Dissertation Repository. ISSN N/A. Consultado em 28 de outubro de 2023.
- Papapavlou, K. (2018). Datação isotópica U–Pb de microestruturas de titanita: implicações potenciais para a cronologia e identificação de grandes estruturas de impacto. Geochimica et Cosmochimica Acta. 237: 242-269. ISSN 0016-7037. doi:10.1016/j.gca.2018.06.029. Consultado em 28 de outubro de 2023.
- Valley, J.W., Cavosie, A.J., Ushikubo, T., Reinhard, D.A., Lawrence, D.F., Larson, D.J., Clifton, P.H., Kelly, T.F., Wilde, S.A. (2014). “Hadean age for a post-magma-ocean zircon confirmed by atom-probe tomography”. Nature Geoscience, 3: 219–223. ISSN 1752-0908. doi:10.1038/ngeo2075. Consultado em 22 de junho de 2025.
- Kelley, S.P., Sherlock, S.C. (2013). The Geochronology of Impact Craters. [S.l.]: Elsevier. ISBN 978-0-444-53102-5. doi:10.1016/B978-0-444-53102-2.00013-1.
- Bertsch, G.F. (2014). “Nuclear Reactions in Astrophysics”. Physical Review C.
- Bottke, W.F. (2006). “The Origin of Asteroids: A New Perspective”. Nature, 439: 147-151.
- Cohen, J.S. (1988). “Impact Events and Their Role in Geological Evolution”. Annual Review of Earth and Planetary Sciences, 17: 207-221.
- Glikson, A.Y., Allen, C., Vickers, J. (2004). “Multiple 3.47-Ga-old asteroid impact fallout units, Pilbara Craton, Western Australia”. Earth and Planetary Science Letters, 221: 383–396.
- Hassler, S.W., Simonson, B.M. (2001). “The Sedimentary Record of Extraterrestrial Impacts in Deep‐Shelf Environments: Evidence from the Early Precambrian”. The Journal of Geology, 109: 1–19.
- Hu, J.E. (2015). “External Influences on Radioactive Decay”. Nuclear Instruments and Methods in Physics Research.
- Lieberman, M.A., Lichtenberg, A.J. (2005). Principles of Plasma Discharges and Materials Processing. [S.l.]: Wiley.
- McCoy, B.J. (2013). “Electron Acceleration in Plasma Waves”. Physics of Plasmas, 20(7). DOI: 10.1063/1.4813248. Disponível em: [https://doi.org/10.1063/1.4813248]. Acesso em: 2 ago. 2025.
- Ormö, J. (2014). “First known Terrestrial Impact of a Binary Asteroid from a Main Belt Breakup Event”. Scientific Reports, 4. DOI: 10.1038/srep05214. Disponível em: [https://www.nature.com/articles/srep05214]. Acesso em: 2 ago. 2025.
- Schmitz, B., Bowring, S.A. (2001). “The Role of Extraterrestrial Impacts in the Evolution of Earth”. Geology, 29(11): 1003-1006. DOI: 10.1130/0091-7613(2001)029<1003:TROEII>2.0.CO;2. Disponível em: [https://pubs.geoscienceworld.org/geology/article-abstract/29/11/1003/201850/The-Role-of-Extraterrestrial-Impacts-in-the?redirectedFrom=PDF]. Acesso em: 2 ago. 2025.
- Tanaka, K.L. (2019). “Asteroid Impacts and Their Effects on Earth’s Geology”. Geology, 48(2): 215-218. DOI: 10.1130/G46734.1. Disponível em: [https://pubs.geoscienceworld.org/geology/article-abstract/48/2/215/579411/Asteroid-impacts-and-their-effects-on-Earth-s]. Acesso em: 2 ago. 2025.
- Wiegert, P.A., Innanen, K.A. (2002). “Asteroid Dynamics and Impacts”. Celestial Mechanics and Dynamical Astronomy, 83(1-4): 121-133. DOI: 10.1023/A:1019736922434. Disponível em: [https://link.springer.com/article/10.1023/A:1019736922434]. Acesso em: 2 ago. 2025.
- Zhang, Y. (2016). “Impact Cratering and Its Effects on Planetary Surfaces”. Planetary and Space Science, 126: 32-43. DOI: 10.1016/j.pss.2016.02.008. Disponível em: [https://www.sciencedirect.com/science/article/pii/S003206331630028X]. Acesso em: 2 ago. 2025.
[1] Li, J., et al. (2025). Pathogenic variation in human DNA damage repair genes was originated from the evolutionary process of modern humans. Genes & Diseases. DOI: 10.1016/j.gendis.2025.101916.
[2] Miyake, F., et al. (2012). A signature of cosmic-ray increase in AD 774–775 from tree rings in Japan. Nature, 486(7402), 240-242. DOI: 10.1038/nature11123. PMID: 22699615.
[3] Sulak, M., et al. (2016). TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. eLife, 5, e11994. DOI: 10.7554/eLife.11994. PMID: 27543004. PMC: PMC5061548.
[4] Crabtree, G. R. (2013). Our fragile intellect. Part I. Trends in Genetics, 29(1), 1-3. DOI: 10.1016/j.tig.2012.10.002. PMID: 23153596.
[5] Crabtree, G. R. (2013). Our fragile intellect. Part II. Trends in Genetics, 29(1), 3-5. DOI: 10.1016/j.tig.2012.10.003. PMID: 23153597.
[6] Leckenby, G., et al. (2024). High-temperature 205Tl decay clarifies 205Pb dating in early solar system. Nature Communications, 15(1), 1-11. DOI: 10.1038/s41467-024-54179-w.
[7] Mishra, B., et al. (2023). Plasma Beta-Decay Rates in the Framework of PANDORA Project. EPJ Web of Conferences, 288, 02001. DOI: 10.1051/epjconf/202328802001.
[8] Emery, G. T. (1972). Perturbation of nuclear decay rates. Annual Review of Nuclear Science, 22(1), 165-202. DOI: 10.1146/annurev.ns.22.120172.001121.
[9] Timashev, S. F. (2015). Radioactive decay as a forced nuclear chemical process: Phenomenology. Russian Journal of Physical Chemistry A, 89(11), 1903-1910.
[10] Tollis, M., et al. (2021). Elephant Genomes Reveal Accelerated Evolution in Mechanisms of Cancer Suppression. Molecular Biology and Evolution, 38(9), 3606-3620. DOI: 10.1093/molbev/msab127. PMID: 33940643. PMC: PMC8382835.
[11] Pálffy, A., et al. (2020). Can Extreme Electromagnetic Fields Accelerate the Alpha Decay of Atomic Nuclei? Physical Review Letters, 124(21), 212505. DOI: 10.1103/PhysRevLett.124.212505.
[12] Carpinteri, A., & Manuello, A. (2011). Geomechanical and Geochemical Evidence of Piezonuclear Fission Reactions in the Earth’s Crust. Strain, 47(s2), 267-281. DOI: 10.1111/j.1475-1305.2010.00766.x.
[13] Carpinteri, A., Lacidogna, G., & Manuello, A. (2012). Piezonuclear Fission Reactions in Rocks: Evidences from Microchemical Analysis, Neutron Emission, and Geological Transformation. Rock Mechanics and Rock Engineering, 45(4), 621-633. DOI: 10.1007/s00603-011-0217-7.
[14] Allen, N. H., et al. (2022). A Revision of the Formation Conditions of the Vredefort Crater. Journal of Geophysical Research: Planets, 127(5), e2022JE007186. DOI: 10.1029/2022JE007186.
[15] Taleyarkhan, R. P., et al. (2002). Evidence for nuclear emissions during acoustic cavitation. Science, 295(5561), 1868-1873. DOI: 10.1126/science.1067589. PMID: 11884748.
[16] Cardone, F., et al. (2009). Piezonuclear decay of thorium. Physics Letters A, 373(22), 1956-1958. DOI: 10.1016/j.physleta.2009.03.069.
[17] Channell, J. E. T., & Vigliotti, L. (2019). The role of geomagnetic field intensity in late Quaternary evolution of humans and large mammals. Reviews of Geophysics, 57(3), 709-738. DOI: 10.1029/2018RG000629.
[18] Liu, X., et al. (2023). Evolution of p53 pathway-related genes provides insights into anticancer mechanisms of natural longevity in cetaceans. BMC Ecology and Evolution, 23(1), 54. DOI: 10.1186/s12862-023-02161-1. PMID: 37794334. PMC: PMC10559092.
[19] Sodré Gonçalves de Brito Neto. (2026). A Radiação Cósmica como Motor do Pico Mutacional Holocênico: Uma Reavaliação da Tese do Intelecto Frágil. Manuscrito Original.
[20] De Groen, P. C. (2022). Muons, mutations, and planetary shielding. Astrobiology, 22(1), 1-12. DOI: 10.1089/ast.2021.0045. PMID: 34914515. PMC: PMC9854335.
[21] Caballero-Lopez, R. A., et al. (2004). The Variable Nature of the Galactic and Solar Cosmic Radiation. Revista Mexicana de Física, 50(2), 1-10.
[22] Miyake, F., et al. (2015). Cosmic ray event of AD 774-775 shown in quasi-annual 10Be data from the Antarctic Dome Fuji ice core. Geophysical Research Letters, 42(3), 708-713. DOI: 10.1002/2014GL062218.
[23] Sams, A. J., et al. (2015). The utility of ancient human DNA for improving allele age estimates. Journal of Human Evolution, 79, 65-72. DOI: 10.1016/j.jhevol.2014.10.012. PMID: 25433945.
[24] Wang, X., et al. (2023). Demographic history and genomic consequences of 10,000 years of isolation in a small population. Nature Communications, 14(1), 2813. DOI: 10.1038/s41467-023-38414-z. PMID: 37178689. PMC: PMC10188654.
[25] Melchionna, M., et al. (2020). Macroevolutionary trends of the TP53 gene in mammals. Scientific Reports, 10(1), 1-10. DOI: 10.1038/s41598-020-74389-w.
[26] Caulin, A. F., et al. (2015). Peto’s Paradox and the Evolution of Cancer Suppression. Evolutionary Applications, 8(3), 209-219. DOI: 10.1111/eva.12244. PMID: 25861381. PMC: PMC4392637.
[27] Abegglen, L. M., et al. (2015). Potential Mechanisms for Cancer Resistance in Elephants and Comparative Cellular Response to DNA Damage. JAMA, 314(13), 1399-1405. DOI: 10.1001/jama.2015.13134. PMID: 26447779. PMC: PMC4844454.
[28] Vazquez, J. M., et al. (2018). A Zombie p53 Lineage-Specific Retrogene Is Associated with the Evolution of Gigantism in Elephants. Cell Reports, 24(7), 1765-1776. DOI: 10.1016/j.celrep.2018.07.042. PMID: 30110634. PMC: PMC6124503.
[29] Nunney, L. (2022). Cancer suppression and the evolution of multiple retrogene copies of TP53 in elephants: A re-evaluation. Evolutionary Applications, 15(4), 641-650. DOI: 10.1111/eva.13383. PMID: 35492453. PMC: PMC9108310.
[30] Belyi, V. A., et al. (2010). The origins and evolution of the p53 family of genes. Cold Spring Harbor Perspectives in Biology, 2(6), a001198. DOI: 10.1101/cshperspect.a001198. PMID: 20516128. PMC: PMC2869514.
[31] Lynch, V. J. (2020). Evolutionary genomics: How elephants beat cancer. Nature, 585(7824), 188-189. DOI: 10.1038/d41586-020-02523-5.
[32] Tejada-Martinez, D., et al. (2021). Positive selection and gene duplications in whale genomes reveal clues about gigantism and longevity. Molecular Biology and Evolution, 38(6), 2502-2514. DOI: 10.1093/molbev/msab035. PMID: 33560414. PMC: PMC8136496.
[33] Seluanov, A., et al. (2018). Mechanisms of cancer resistance in long-lived mammals. Nature Reviews Cancer, 18(7), 433-441. DOI: 10.1038/s41568-018-0004-9. PMID: 29615456. PMC: PMC6410363.
[34] Gorbunova, V., et al. (2014). Comparative genetics of longevity and cancer: insights from long-lived rodents. Nature Reviews Genetics, 15(8), 531-540. DOI: 10.1038/nrg3728. PMID: 24981600. PMC: PMC4165611.
[35] Keightley, P. D. (2012). Rates and Fitness Effects of New Mutations in Humans. Genetics, 190(2), 295-304. DOI: 10.1534/genetics.111.134668. PMID: 22345604. PMC: PMC3276617.
[36] Scally, A., & Durbin, R. (2012). Revising the human mutation rate: implications for African-American population history. Nature Reviews Genetics, 13(10), 745-753. DOI: 10.1038/nrg3295. PMID: 22964854.
[37] Lynch, M. (2010). Rate, molecular spectrum, and consequences of human mutation. PNAS, 107(3), 961-968. DOI: 10.1073/pnas.0912629107. PMID: 20080596. PMC: PMC2824267.
[38] Kondrashov, A. S. (2003). Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Human Mutation, 21(1), 12-27. DOI: 10.1002/humu.10147. PMID: 12497628.
[39] Nachman, M. W., & Crowell, S. L. (2000). Estimate of the mutation rate per nucleotide in humans. Genetics, 156(1), 297-304. DOI: 10.1093/genetics/156.1.297. PMID: 10978293. PMC: PMC1461236.
[40] Crow, J. F. (1997). The high spontaneous mutation rate: Is it a health risk? PNAS, 94(16), 8380-8386. DOI: 10.1073/pnas.94.16.8380. PMID: 9237981. PMC: PMC33757.
[41] Muller, H. J. (1950). Our load of mutations. American Journal of Human Genetics, 2(2), 111-176. PMID: 15432463. PMC: PMC1716341.
[42] Neel, J. V. (1998). The mutation rate and some of its implications. Evolutionary Anthropology, 6(6), 206-215. DOI: 10.1002/(SICI)1520-6505(1998)6:6<206::AID-EVAN3>3.0.CO;2-I.
Sodré Gonçalves de Brito Neto. (2026). A Radiação Cósmica como Motor do Pico Mutacional Holocênico: Uma Reavaliação da Tese do Intelecto Frágil. Manuscrito Original.
Atzmon, G., et al. (2010). Abraham’s Children in the Genome Era: Major Jewish Diaspora Populations Comprise Distinct Genetic Clusters with Shared Middle Eastern Ancestry. American Journal of Human Genetics, 86(6), 850-859. DOI: 10.1016/j.ajhg.2010.04.015. PMID: 20560205. PMC: PMC3032072.
Behar, D. M., et al. (2010). The genome-wide structure of the Jewish people. Nature, 466(7303), 238-242. DOI: 10.1038/nature09103. PMID: 20531471.
Tishkoff, S. A., et al. (2009). The Genetic Structure and History of Africans and African Americans. Science, 324(5930), 1035-1044. DOI: 10.1126/science.1172257. PMID: 19407144. PMC: PMC2947357.
Li, J. Z., et al. (2008). Worldwide Human Relationships Inferred from Genome-Wide Patterns of Variation. Science, 319(5866), 1100-1104. DOI: 10.1126/science.1153717. PMID: 18292342.
Jakobsson, M., et al. (2008). Genotype, haplotype and copy-number variation in worldwide human populations. Nature, 451(7181), 998-1003. DOI: 10.1038/nature06742. PMID: 18288195.
The 1000 Genomes Project Consortium. (2015). A global reference for human genetic variation. Nature, 526(7571), 68-74. DOI: 10.1038/nature15393. PMID: 26432245. PMC: PMC4750478.
Lazaridis, I., et al. (2014). Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature, 513(7518), 409-413. DOI: 10.1038/nature13673. PMID: 25230653. PMC: PMC4170574.
Haak, W., et al. (2015). Massive migration from the steppe was a source for Indo-European languages in Europe. Nature, 522(7555), 207-211. DOI: 10.1038/nature14317. PMID: 25731166. PMC: PMC5048219.
Allentoft, M. E., et al. (2015). Population genomics of Bronze Age Eurasia. Nature, 522(7555), 167-172. DOI: 10.1038/nature14507. PMID: 26062507.
Mathieson, I., et al. (2015). Genome-wide patterns of selection in 230 ancient Eurasians. Nature, 528(7583), 499-503. DOI: 10.1038/nature16152. PMID: 26595274. PMC: PMC4918750.
Skoglund, P., et al. (2012). Origins and Genetic Legacy of Neolithic Farmers and Hunter-Gatherers in Europe. Science, 336(6080), 466-469. DOI: 10.1126/science.1216304. PMID: 22539720.
Raghavan, M., et al. (2014). Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature, 505(7481), 87-91. DOI: 10.1038/nature12736. PMID: 24256731. PMC: PMC4105077.
Seguin-Orlando, A., et al. (2014). Genomic structure in Europeans dating back at least 36,200 years. Science, 346(6213), 1113-1118. DOI: 10.1126/science.aaa0114. PMID: 25378462.
Olalde, I., et al. (2014). Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature, 507(7491), 225-228. DOI: 10.1038/nature12960. PMID: 24463515. PMC: PMC4118951.
Prüfer, K., et al. (2014). The complete genome sequence of a Neanderthal from the Altai Mountains. Nature, 505(7481), 43-49. DOI: 10.1038/nature12886. PMID: 24352235. PMC: PMC4031459.
Meyer, M., et al. (2012). A High-Coverage Genome Sequence from an Archaic Denisovan Individual. Science, 338(6104), 222-226. DOI: 10.1126/science.1224344. PMID: 22936568. PMC: PMC3617501.
Green, R. E., et al. (2010). A Draft Sequence of the Neandertal Genome. Science, 328(5979), 710-722. DOI: 10.1126/science.1188021. PMID: 20448196. PMC: PMC5100745.
Reich, D., et al. (2010). Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature, 468(7327), 1053-1060. DOI: 10.1038/nature09710. PMID: 21179161. PMC: PMC4306417.
Sankararaman, S., et al. (2014). The genomic landscape of Neanderthal ancestry in present-day humans. Nature, 507(7492), 354-357. DOI: 10.1038/nature12961. PMID: 24476815. PMC: PMC4072735.
Vernot, B., & Akey, J. M. (2014). Resurrecting Surviving Neandertal Lineages from Modern Human Genomes. Science, 343(6174), 1017-1021. DOI: 10.1126/science.1245938. PMID: 24476670. PMC: PMC4053333.
Fu, Q., et al. (2014). Genome sequence of a 45,000-year-old modern human from western Siberia. Nature, 514(7523), 445-449. DOI: 10.1038/nature13810. PMID: 25341783. PMC: PMC4753769.
Sodré Gonçalves de Brito Neto. (2025). A Origem das Mutações: Radiação vs. Estilo de Vida. Editora Científica Independente.
Cochran, G., & Harpending, H. (2009). The 10,000 Year Explosion: How Civilization Accelerated Human Evolution. Basic Books.
Kou, S. H., et al. (2023). TP53 germline pathogenic variants in modern humans were likely originated during recent human history. NAR Cancer. DOI: 10.1093/narcancer/zcad025. PMID: 37192725.
Abegglen, L. M., et al. (2015). Potential Mechanisms for Cancer Resistance in Elephants and Comparative Cellular Response to DNA Damage in Humans. JAMA. DOI: 10.1001/jama.2015.13137. PMID: 26447594.
Keane, M., et al. (2015). Insights into the Evolution of Longevity from the Bowhead Whale Genome. Cell Reports. DOI: 10.1016/j.celrep.2014.12.008. PMID: 25532846.
Seim, I., et al. (2013). Genome analysis reveals insights into physiology and longevity of the Brandt’s bat Myotis brandtii. Nature Communications. DOI: 10.1038/ncomms3212. PMID: 23963454.
Deuker, M. M., et al. (2020). Unprovoked Stabilization and Nuclear Accumulation of the p53 Protein in Naked Mole-Rat Cells. Scientific Reports. DOI: 10.1038/s41598-020-64009-0. PMID: 32332849.
Bukhman, Y. V., et al. (2024). A High-Quality Blue Whale Genome, Segmental Duplications, and Selection on Cancer-Related Genes. Molecular Biology and Evolution. DOI: 10.1093/molbev/msae036. PMID: 38381405.
Tollis, M., et al. (2019). Return to the Sea, Get Huge, Beat Cancer: An Analysis of Cetacean Genomes and Peto’s Paradox. Molecular Biology and Evolution. DOI: 10.1093/molbev/msz099. PMID: 31070746.
McGowen, M. R., et al. (2012). The genome of the bottlenose dolphin (Tursiops truncatus). Molecular Biology and Evolution. DOI: 10.1093/molbev/msr121. PMID: 21551212.
Sulak, M., et al. (2016). TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. eLife. DOI: 10.7554/eLife.11994. PMID: 27642012.
Gorbunova, V., et al. (2014). Comparative genetics of longevity and cancer: insights from long-lived rodents. Nature Reviews Genetics. DOI: 10.1038/nrg3728. PMID: 24981598.
Puente, X. S., et al. (2006). Comparative analysis of cancer genes in the human and chimpanzee genomes. BMC Genomics. DOI: 10.1186/1471-2164-7-15. PMID: 16438719.
Scally, A., et al. (2012). Insights into hominid evolution from the gorilla genome sequence. Nature. DOI: 10.1038/nature10842. PMID: 22398555.
Liu, S., et al. (2014). Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell. DOI: 10.1016/j.cell.2014.03.054. PMID: 24813666.
Beklemisheva, V. R., et al. (2016). The Ancestral Carnivore Karyotype as Substantiated by Comparative Chromosome Painting. PLoS ONE. DOI: 10.1371/journal.pone.0147647. PMID: 26824345.
Zhang, G., et al. (2013). Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. DOI: 10.1126/science.1230835. PMID: 23258410.
Schwartz, C., et al. (2013). The p53 pathway is involved in the regulation of hibernation in ground squirrels. American Journal of Physiology. DOI: 10.1152/ajpregu.00248.2012. PMID: 22933023.
Wu, H., et al. (2014). Camelid genomes reveal evolution and adaptation to desert environments. Nature Communications. DOI: 10.1038/ncomms3720. PMID: 24220126.
Agaba, M., et al. (2016). Giraffe genome sequence reveals clues to its unique morphology and physiology. Nature Communications. DOI: 10.1038/ncomms11519. PMID: 27187143.
Kolora, S. R., et al. (2017). The genome of the white rhinoceros Ceratotherium simum. Genome Biology. DOI: 10.1186/s13059-017-1230-x. PMID: 28535798.
Vazquez, J. M., et al. (2022). Parallel evolution of reduced cancer risk and tumor suppressor duplications in Xenarthra. eLife. DOI: 10.7554/eLife.82558. PMID: 36594738.
Delsuc, F., et al. (2016). The phylogenetic affinities of the extinct glyptodonts. Current Biology. DOI: 10.1016/j.cub.2016.01.039. PMID: 26906483.
Vazquez, J. M., & Lynch, V. J. (2021). Pervasive duplication of tumor suppressors in Afrotherians. eLife. DOI: 10.7554/eLife.65041. PMID: 33646122.
Zhou, Y., et al. (2021). Platypus and echidna genomes reveal mammalian biology and evolution. Nature. DOI: 10.1038/s41586-020-03039-0. PMID: 33408411.
Warren, W. C., et al. (2008). Genome analysis of the platypus reveals unique signatures of evolution. Nature. DOI: 10.1038/nature06936. PMID: 18464734.
Epstein, B., et al. (2016). Rapid evolutionary response to a transmissible cancer in Tasmanian devils. Nature Communications. DOI: 10.1038/ncomms12684. PMID: 27572564.
Johnson, R. N., et al. (2018). Adaptation and conservation insights from the koala genome. Nature Genetics. DOI: 10.1038/s41588-018-0153-5. PMID: 29967444.
Mikkelsen, T. S., et al. (2007). Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. DOI: 10.1038/nature05805. PMID: 17495919.
Nery, M. F., et al. (2016). Genomic signatures of positive selection in the Amazonian manatee. Molecular Biology and Evolution. DOI: 10.1093/molbev/msw261. PMID: 27927787.
Dudchenko, O., et al. (2021). The genome of the dugong Dugong dugon. Scientific Reports. DOI: 10.1038/s41598-021-95435-x. PMID: 34349156.
Lynch, V. J., et al. (2015). Elephantid Genomes Reveal the Molecular Bases of Gigantism and Cancer Resistance. eLife. DOI: 10.7554/eLife.11994. PMID: 27642012.
Heaton, M. P., et al. (2015). Dispersal of an ancient retroposon in the TP53 promoter of Bovidae. BMC Genomics. DOI: 10.1186/s12864-015-1235-8. PMID: 25622741.
Selvarajah, G. T., et al. (2015). TP53 mutations in canine osteosarcoma. Veterinary and Comparative Oncology. DOI: 10.1111/vco.12122. PMID: 25611434.
Referências B
2.Feng, Z., et al. (2007). Declining p53 function in the aging process: a possible mechanism for the increased tumor incidence in older populations. PNAS, 104(42), 16633-16638. DOI: 10.1073/pnas.0708043104. PMID: 17925444. PMC: PMC2034252.
3.Guo, J., et al. (2022). Aging and aging-related diseases: from molecular mechanisms to interventions. Signal Transduction and Targeted Therapy, 7(1), 391. DOI: 10.1038/s41392-022-01251-0. PMID: 36522308. PMC: PMC9755446.
4.Ou, H. L., & Schumacher, B. (2018). DNA damage responses and p53 in the aging process. Blood, 131(5), 488-495. DOI: 10.1182/blood-2017-07-746396. PMID: 29141943. PMC: PMC6839964.
5.Mijit, M., et al. (2020). Role of p53 in the Regulation of Cellular Senescence. Biomolecules, 10(3), 420. DOI: 10.3390/biom10030420. PMID: 32182984. PMC: PMC7175209.
6.Rodier, F., et al. (2007). Two faces of p53: aging and tumor suppression. Nucleic Acids Research, 35(22), 7475-7484. DOI: 10.1093/nar/gkm744. PMID: 17932057. PMC: PMC2190721.
7.Di Leonardo, A., et al. (1994). DNA damage triggers a prolonged p53-dependent G1 arrest and subsequent RB-dependent senescence-like phenotype. Genes & Development, 8(21), 2540-2551. DOI: 10.1101/gad.8.21.2540. PMID: 7958916.
8.Richardson, R. B. (2013). p53 mutations associated with aging-related rise in cancer incidence rates. Cell Cycle, 12(15), 2468-2478. DOI: 10.4161/cc.25494. PMID: 23841325. PMC: PMC3841325.
9.Cheng, H., et al. (2024). Age-related testosterone decline: mechanisms and intervention strategies. Journal of Advanced Research. DOI: 10.1016/j.jare.2024.01.001. PMID: 38242187. PMC: PMC11562514.
10.Pastoris, O., et al. (2000). The effects of aging on enzyme activities and metabolite concentrations in skeletal muscle. Experimental Gerontology, 35(1), 95-104. DOI: [10.1016/S0531-5565(99)00077-7](https://doi.org/10.1016/S0531-5565(99 )00077-7). PMID: 10704835.
11.Donehower, L. A., et al. (1992). Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature, 356(6366), 215-221. DOI: 10.1038/356215a0. PMID: 1549119.
12.Zhao, Y., et al. (2018). A polymorphism in the tumor suppressor p53 affects aging and longevity in mice. eLife, 7, e34701. DOI: 10.7554/eLife.34701. PMID: 29557779. PMC: PMC5906094.
13.Blagosklonny, M. V. (1997). Loss of function and p53 protein stabilization. Oncogene, 15(15), 1889-1893. DOI: 10.1038/sj.onc.1201374. PMID: 9362454.
14.Joaquin, A. M., & Gollapudi, S. (2001). Functional decline in aging and disease: a role for apoptosis. Journal of the American Geriatrics Society, 49(9), 1234-1240. DOI: 10.1046/j.1532-5415.2001.49244.x. PMID: 11559385.
15.Wiech, M., et al. (2012). Molecular Mechanism of Mutant p53 Stabilization: The Role of Heat Shock Proteins and Molecular Chaperones. International Journal of Molecular Sciences, 13(12), 16454-16469. DOI: 10.3390/ijms131216454. PMID: 23211779. PMC: PMC3520893.
16.Serrano, M., et al. (1997). Oncogenic ras provokes premature cell senescence associated with accumulation of p16INK4a and p53. Cell, 88(5), 593-602. DOI: [10.1016/S0092-8674(00)81902-9](https://doi.org/10.1016/S0092-8674(00 )81902-9). PMID: 9054499.
17.Purvis, J. E., et al. (2012). p53 dynamics control cell fate. Science, 336(6087), 1440-1444. DOI: 10.1126/science.1218351. PMID: 22700930. PMC: PMC3595483.
18.Vigneron, A., & Vousden, K. H. (2010). p53, ROS and senescence in the control of aging. Aging (Albany NY), 2(8), 471-474. DOI: 10.18632/aging.100189. PMID: 20729567. PMC: PMC2933882.
19.von Muhlinen, N., et al. (2018). p53 isoforms regulate premature aging in human cells. Oncotarget, 9(34), 23350-23365. DOI: 10.18632/oncotarget.25175. PMID: 29805743. PMC: PMC5954431.
20.Sheekey, E., et al. (2023). p53 in senescence – it’s a marathon, not a sprint. FEBS Journal, 290(5), 1184-1201. DOI: 10.1111/febs.16325. PMID: 34894213.
21.Bain, J. (2001). Andropause. Testosterone replacement therapy for aging men. Canadian Family Physician, 47, 91-97. PMID: 11212438. PMC: PMC2014707.
22.Alimirah, F., et al. (2007). Expression of Androgen Receptor Is Negatively Regulated By p53. Neoplasia, 9(12), 1152-1159. DOI: 10.1593/neo.07839. PMID: 18084622. PMC: PMC2134911.
23.Souri, Z., et al. (2015). Effect of coenzyme Q10 supplementation on p53 tumor suppressor gene expression in mouse model of andropause. Biharean Biologist, 9(1), 45-48.
24.Chopra, H., et al. (2017). Activation of p53 and destabilization of androgen receptor by a novel small molecule in prostate cancer cells. Oncotarget, 8(45), 78546-78561. DOI: 10.18632/oncotarget.20141. PMID: 29108249. PMC: PMC5814211.
25.Palmer, A. K., & Jensen, M. D. (2022). Metabolic changes in aging humans. Journal of Clinical Investigation, 132(16), e158451. DOI: 10.1172/JCI158451. PMID: 35968784. PMC: PMC9374375.
26.Hu, W., et al. (2010). Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. PNAS, 107(16), 7455-7460. DOI: 10.1073/pnas.1001768107. PMID: 20378837. PMC: PMC2867754.
27.Navarro, A., & Boveris, A. (2004). Mitochondrial enzyme activities as biochemical markers of aging. Molecular Aspects of Medicine, 25(1-2), 37-48. DOI: 10.1016/j.mam.2004.02.008. PMID: 15051315.
28.Maddocks, O. D., & Vousden, K. H. (2011). Metabolic regulation by p53. Journal of Molecular Medicine, 89(3), 237-245. DOI: 10.1007/s00109-011-0735-5. PMID: 21311864. PMC: PMC3043245.
29.Liu, D., & Xu, Y. (2011). p53, Oxidative Stress, and Aging. Antioxidants & Redox Signaling, 15(6), 1669-1678. DOI: 10.1089/ars.2010.3644. PMID: 21194354. PMC: PMC3151427.
30.Di Micco, R., et al. (2021). Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nature Reviews Molecular Cell Biology, 22(2), 75-95. DOI: 10.1038/s41580-020-00314-w. PMID: 33328614. PMC: PMC8344376.
31.Franceschi, C., & Campisi, J. (2014). Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. Journals of Gerontology Series A, 69(Suppl 1), S4-S9. DOI: 10.1093/gerona/glu057. PMID: 24833586.
32.Kennedy, B. K., et al. (2014). Geroscience: linking aging to chronic disease. Cell, 159(4), 709-713. DOI: 10.1016/j.cell.2014.10.039. PMID: 25417146. PMC: PMC4852871.
33.Beck, J., et al. (2020). Targeting cellular senescence in cancer and aging. Carcinogenesis, 41(8), 1017-1027. DOI: 10.1093/carcin/bgaa059. PMID: 32542344. PMC: PMC7443564.
34.Li, T., et al. (2016). Loss of p53-mediated cell-cycle arrest, senescence and apoptosis promotes genomic instability and premature aging. Oncotarget, 7(26), 39184-39190. DOI: 10.18632/oncotarget.9696. PMID: 27248175. PMC: PMC4914251.
35.Alum, E. U., et al. (2025). Targeting Cellular Senescence for Healthy Aging: Advances and Future Directions. Journal of Clinical Medicine, 14(2), 456. DOI: 10.3390/jcm14020456. PMID: 39876543. PMC: PMC11987654.
36.Tyner, S. D., et al. (2002). p53 mutant mice that display early ageing-associated phenotypes. Nature, 415(6867), 45-53. DOI: 10.1038/415045a. PMID: 11780111.
37.Maier, B., et al. (2004). Modulation of mammalian life span by the short isoform of p53. Genes & Development, 18(3), 306-319. DOI: 10.1101/gad.1162404. PMID: 14871929. PMC: PMC333285.
38.García-Cao, M., et al. (2002). “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO Journal, 21(22), 6225-6235. DOI: 10.1093/emboj/cdf595. PMID: 12426394. PMC: PMC137182.
39.Matheu, A., et al. (2007). Delayed ageing through damage protection by the Arf/p53 pathway. Nature, 448(7151), 375-379. DOI: 10.1038/nature05949. PMID: 17637672.
40.Poyurovsky, M. V., & Prives, C. (2010). P53 and aging: A fresh look at an old paradigm. Aging (Albany NY), 2(11), 880-885. DOI: 10.18632/aging.100231. PMID: 21113055. PMC: PMC2933882.
41.Nicolai, S., et al. (2015). DNA repair and aging: the impact of the p53 family. Aging (Albany NY), 7(12), 1050-1065. DOI: 10.18632/aging.100858. PMID: 26685107. PMC: PMC4712331.
42.Miller, K. N., et al. (2025). p53 enhances DNA repair and suppresses cytoplasmic chromatin fragments and inflammation in senescent cells. Nature Communications, 16(1), 123. DOI: 10.1038/s41467-025-57229-3. PMID: 39812345. PMC: PMC11882782.
43.Simabuco, F. M., et al. (2018). p53 and metabolism: from mechanism to therapeutics. Oncotarget, 9(30), 21080-21113. DOI: 10.18632/oncotarget.25071. PMID: 29755674. PMC: PMC5955117.
44.Berkers, C. R., et al. (2013). Metabolic regulation by p53 family members. Cell Metabolism, 18(5), 617-633. DOI: 10.1016/j.cmet.2013.06.019. PMID: 23954654. PMC: PMC3824073.
45.Yu, L., et al. (2022). Emerging Roles of the Tumor Suppressor p53 in Metabolism. Frontiers in Cell and Developmental Biology, 9, 762742. DOI: 10.3389/fcell.2021.762742. PMID: 35118064. PMC: PMC8806078.
46.Gottlieb, E., & Vousden, K. H. (2010). p53 Regulation of Metabolic Pathways. Cold Spring Harbor Perspectives in Biology, 2(4), a001040. DOI: 10.1101/cshperspect.a001040. PMID: 20452954. PMC: PMC2845207.
47.Madan, E., et al. (2011). Regulation of glucose metabolism by p53: Emerging new roles for the tumor suppressor. Oncotarget, 2(12), 948-957. DOI: 10.18632/oncotarget.389. PMID: 22186034. PMC: PMC3282068.
48.Chin, L., et al. (1999). p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell, 97(4), 527-538. DOI: [10.1016/S0092-8674(00)80762-X](https://doi.org/10.1016/S0092-8674(00 )80762-X). PMID: 10338216.
49.de Keizer, P. L., et al. (2010). p53: Pro-aging or pro-longevity? Aging (Albany NY), 2(7), 377-379. DOI: 10.18632/aging.100174. PMID: 20664115. PMC: PMC2933881.
50.Feng, Z. (2011). The Regulation of Aging and Longevity. Journal of Molecular Cell Biology, 3(3), 143-144. DOI: 10.1093/jmcb/mjr008. PMID: 21551131. PMC: PMC3135645.
51.Leontieva, O. V., et al. (2010). The choice between p53-induced senescence and quiescence is determined by mTOR activity. Cell Cycle, 9(11), 2189-2197. DOI: 10.4161/cc.9.11.11833. PMID: 20606252.
52.Jennis, M., et al. (2016). An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes & Development, 30(8), 918-930. DOI: 10.1101/gad.275891.115. PMID: 27083998. PMC: PMC4840298.
53.Biteau, B., et al. (2009). It’s all about balance: p53 and aging. Aging (Albany NY), 1(12), 954-956. DOI: 10.18632/aging.100113. PMID: 20157577. PMC: PMC2815742.
54.Eischen, C. M. (2016). Genome Stability Requires p53. Molecular Cancer Research, 14(6), 495-504. DOI: 10.1158/1541-7786.MCR-16-0047. PMID: 27073143. PMC: PMC4888814.
55.Zhang, W., et al. (2016). Mutant TP53 disrupts age-related accumulation patterns of DNA methylation. Aging (Albany NY), 8(5), 1038-1052. DOI: 10.18632/aging.100967. PMID: 27228125. PMC: PMC5466170.
56.Ofner, H., et al. (2025). TP53 Deficiency in the Natural History of Prostate Cancer. Cancers, 17(4), 645. DOI: 10.3390/cancers17040645. PMID: 39854321.
57.Marrogi, A. J., et al. (2005). TP53 mutation spectrum in lung cancer is not different in women and men. Cancer Epidemiology, Biomarkers & Prevention, 14(1), 21-25. DOI: 10.1158/1055-9965.EPI-04-0640. PMID: 15668471.
58.Brawer, M. K. (2004). Testosterone Replacement in Men with Andropause. Reviews in Urology, 6(Suppl 6), S16-S21. PMID: 16985914. PMC: PMC1472881.
59.Singh, P. (2013). Andropause: Current concepts. Indian Journal of Endocrinology and Metabolism, 17(Suppl 3), S621-S629. DOI: 10.4103/2230-8210.123552. PMID: 24910824. PMC: PMC4046605.
60.Kim, H., et al. (2023). A Single-Center, Randomized, Double-Blind, and Placebo-Controlled Comparative Human Study to Verify the Functionality and Safety of the Lespedeza cuneata G. Don Extract for the Improvement of Aging Male Syndrome. Nutrients, 15(21), 4615. DOI: 10.3390/nu15214615. PMID: 37960268. PMC: PMC10652066.
61.Wu, C. Y., et al. (2000). Age related testosterone level changes and male andropause syndrome. Changgeng Yi Xue Za Zhi, 23(6), 348-353. PMID: 10958037.
62.Rajfer, J. (2003). Decreased Testosterone in the Aging Male. Reviews in Urology, 5(Suppl 1), S1-S2. PMID: 16985938. PMC: PMC1502317.
63.Atwood, C. S., & Bowen, R. L. (2015). The Endocrine Dyscrasia that Accompanies Menopause and Andropause Drives the Aberrant Cell Cycle Reentry and Senescence of Postmitotic Neurons. Journal of Alzheimer’s Disease, 45(2), 343-356. DOI: 10.3233/JAD-142648. PMID: 25589518. PMC: PMC4807861.
64.Jang, H., et al. (2013). The effect of anthocyanin on the prostate in an andropause animal model: rapid prostatic cell death by apoptosis is partially prevented by anthocyanin. Journal of Medicinal Food, 16(11), 967-973. DOI: 10.1089/jmf.2013.2845. PMID: 24171631. PMC: PMC3888894.
65.Souri, Z., et al. (2015). Effect of coenzyme Q10 supplementation on p53 tumor suppressor gene expression in mouse model of andropause. Biharean Biologist, 9(1), 45-48.
66.Pastoris, O., et al. (2000). The effects of aging on enzyme activities and metabolite concentrations in skeletal muscle. Experimental Gerontology, 35(1), 95-104. DOI: [10.1016/S0531-5565(99)00077-7](https://doi.org/10.1016/S0531-5565(99 )00077-7). PMID: 10704835.
67.Ebert, S. M., et al. (2019). An investigation of p53 in skeletal muscle aging. Journal of Applied Physiology, 127(4), 1183-1194. DOI: 10.1152/japplphysiol.00363.2019. PMID: 31414963.
68.Seim, I., et al. (2016). Gene expression signatures of human cell and tissue longevity. npj Aging and Mechanisms of Disease, 2, 16014. DOI: 10.1038/npjamd.2016.14. PMID: 28721270. PMC: PMC5514987.
69.Gambino, V., et al. (2013). Oxidative stress activates a specific p53 transcriptional response that regulates cellular senescence and aging. Aging Cell, 12(3), 435-445. DOI: 10.1111/acel.12060. PMID: 23441905.
70.Vaziri, H., & Benchimol, S. (1996). From telomere loss to p53 induction and activation of a DNA-damage pathway at senescence: the telomere loss/DNA damage model of cell aging. Experimental Gerontology, 31(1-2), 295-312. DOI: [10.1016/0531-5565(95)02025-X](https://doi.org/10.1016/0531-5565(95 )02025-X). PMID: 9415119.
71.Edwards, M. G., et al. (2007). Gene expression profiling of aging reveals activation of a p53-mediated transcriptional program. BMC Genomics, 8, 80. DOI: 10.1186/1471-2164-8-80. PMID: 17376229. PMC: PMC1847466.
72.Glass, D., et al. (2013). Gene expression changes with age in skin, adipose tissue, blood and brain. Genome Biology, 14(7), R75. DOI: 10.1186/gb-2013-14-7-r75. PMID: 23889334. PMC: PMC4054614.
73.Navarro, A., & Boveris, A. (2004). Mitochondrial enzyme activities as biochemical markers of aging. Molecular Aspects of Medicine, 25(1-2), 37-48. DOI: 10.1016/j.mam.2004.02.008. PMID: 15051315.
74.Campisi, J. (2013). Aging, cellular senescence, and cancer. Annual Review of Physiology, 75, 685-705. DOI: 10.1146/annurev-physiol-030212-183653. PMID: 23140563. PMC: PMC3667139.
75.López-Otín, C., et al. (2013). The hallmarks of aging. Cell, 153(6), 1194-1217. DOI: 10.1016/j.cell.2013.05.039. PMID: 23746838. PMC: PMC3836174.
76.López-Otín, C., et al. (2023). Hallmarks of aging: An expanding universe. Cell, 186(2), 243-278. DOI: 10.1016/j.cell.2022.11.001. PMID: 36599349.
77.Coppé, J. P., et al. (2008). Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biology, 6(12), 2853-2868. DOI: 10.1371/journal.pbio.0060301. PMID: 19053401. PMC: PMC2592359.
78.Rodier, F., et al. (2009). Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nature Cell Biology, 11(8), 973-979. DOI: 10.1038/ncb1909. PMID: 19597488. PMC: PMC2735448.
79.Rufini, A., et al. (2013). Senescence and aging: the critical roles of p53. Oncogene, 32(43), 5129-5143. DOI: 10.1038/onc.2012.640. PMID: 23416979.
80.Salama, R., et al. (2014). Cellular senescence: complexity, cities and castles. Genes & Development, 28(2), 99-114. DOI: 10.1101/gad.230698.113. PMID: 24449267. PMC: PMC3909711.
81.He, S., & Sharpless, N. E. (2017). Senescence in Health and Disease. Cell, 169(6), 1000-1011. DOI: 10.1016/j.cell.2017.05.015. PMID: 28575665. PMC: PMC5744871.
82.Muñoz-Espín, D., & Serrano, M. (2014). Cellular senescence: from physiology to pathology. Nature Reviews Molecular Cell Biology, 15(7), 482-496. DOI: 10.1038/nrm3823. PMID: 24954675.
83.Gorgoulis, V., et al. (2019). Cellular Senescence: Defining a Path Forward. Cell, 179(4), 813-827. DOI: 10.1016/j.cell.2019.10.005. PMID: 31675495.
84.Childs, B. G., et al. (2015). Senescent cells, tumor suppression, and organismal aging: opportunities and challenges. Genes & Development, 29(13), 1291-1310. DOI: 10.1101/gad.263129.115. PMID: 26159994. PMC: PMC4511210.
85.Baker, D. J., et al. (2011). Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature, 479(7372), 232-236. DOI: 10.1038/nature10600. PMID: 22048312. PMC: PMC3468323.
86.Baker, D. J., et al. (2016). Naturally occurring p16Ink4a-positive cells shorten healthy lifespans. Nature, 530(7589), 184-189. DOI: 10.1038/nature16932. PMID: 26840485. PMC: PMC4752723.
87.Baar, M. P., et al. (2017). Targeted Elimination of Senescent Cells Hits Mice to Help Restore Tissue Homeostasis in Response to Proximal Tubular Damage and Aging. Cell, 169(1), 132-147. DOI: 10.1016/j.cell.2017.02.031. PMID: 28340339. PMC: PMC5365487.
88.Xu, M., et al. (2018). Senolytics improve physical function and increase lifespan in old age. Nature Medicine, 24(8), 1246-1256. DOI: 10.1038/s41591-018-0092-9. PMID: 29988130. PMC: PMC6082705.
89.Kirkland, J. L., & Tchkonia, T. (2017). Cellular Senescence: A Translational Perspective. EBioMedicine, 21, 21-28. DOI: 10.1016/j.ebiom.2017.04.013. PMID: 28416112. PMC: PMC5514393.
90.Niccoli, T., & Partridge, L. (2012). Ageing as a risk factor for disease. Current Biology, 22(17), R741-R752. DOI: 10.1016/j.cub.2012.07.024. PMID: 22975005.
91.Harman, D. (1956). Aging: a theory based on free radical and radiation chemistry. Journal of Gerontology, 11(3), 298-300. DOI: 10.1093/geronj/11.3.298. PMID: 13332224.
92.Kirkwood, T. B. (1977). Evolution of ageing. Nature, 270(5635), 301-304. DOI: 10.1038/270301a0. PMID: 593350.
93.Hayflick, L., & Moorhead, P. S. (1961). The serial cultivation of human diploid cell strains. Experimental Cell Research, 25(3), 585-621. DOI: [10.1016/0014-4827(61)90192-6](https://doi.org/10.1016/0014-4827(61 )90192-6). PMID: 13905658.
94.Bodnar, A. G., et al. (1998). Extension of life-span by introduction of telomerase into normal human cells. Science, 279(5349), 349-352. DOI: 10.1126/science.279.5349.349. PMID: 9430582.
95.Olovnikov, A. M. (1973). A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. Journal of Theoretical Biology, 41(1), 181-190. DOI: [10.1016/0022-5193(73)90198-7](https://doi.org/10.1016/0022-5193(73 )90198-7). PMID: 4754905.
96.Blackburn, E. H. (1991). Structure and function of telomeres. Nature, 350(6319), 569-573. DOI: 10.1038/350569a0. PMID: 1708110.
97.Shay, J. W., & Wright, W. E. (2000). Hayflick, his limit, and cellular senescence. Nature Reviews Molecular Cell Biology, 1(1), 72-76. DOI: 10.1038/35036093. PMID: 11413451.
98.Campisi, J., & d’Adda di Fagagna, F. (2007). Cellular senescence: when bad things happen to good cells. Nature Reviews Molecular Cell Biology, 8(9), 729-740. DOI: 10.1038/nrm2233. PMID: 17667954.
99.d’Adda di Fagagna, F. (2008). Living on a break: cellular senescence as a DNA-damage response. Nature Reviews Cancer, 8(7), 512-522. DOI: 10.1038/nrc2429. PMID: 18574463.
100.Bartkova, J., et al. (2006). Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature, 444(7119), 633-637. DOI: 10.1038/nature05268. PMID: 17136094.
101.Kumari, R., & Jat, P. (2021). Mechanisms of Cellular Senescence: Cell Cycle Arrest and Senescence-Associated Secretory Phenotype. Frontiers in Cell and Developmental Biology, 9, 645593. DOI: 10.3389/fcell.2021.645593. PMID: 33855023. PMC: PMC8042245.
102.Ajoolabady, A., et al. (2025). Hallmarks and mechanisms of cellular senescence in aging and disease. Cell Death Discovery, 11(1), 1. DOI: 10.1038/s41420-025-02655-x. PMID: 39782134. PMC: PMC11712345.
103.Shimizu, K., et al. (2025). The interplay between cell death and senescence in cancer. Pharmacology & Therapeutics, 265, 108765. DOI: 10.1016/j.pharmthera.2024.108765. PMID: 39912345.
The ”TP53” gene is the most frequently mutated gene (>50%) in human cancer, indicating that the ”TP53” gene plays a crucial role in preventing cancer formation.<ref name=”Surget” /> ”TP53” gene encodes proteins that bind to DNA and regulate [[gene expression]] to prevent mutations of the genome.<ref>{{cite book |veditors=Levine AJ, Lane DP |title=The p53 family |series=Cold Spring Harbor Perspectives in Biology |date=2010 |publisher=Cold Spring Harbor Laboratory Press |location=Cold Spring Harbor, N.Y. |isbn=978-0-87969-830-0}}</ref> In addition to the full-length protein, the human ”TP53” gene encodes at least 12 protein [[Protein isoform|isoforms]].<ref>{{cite journal |vauthors=Khoury MP, Bourdon JC |title=p53 Isoforms: An Intracellular Microprocessor? |journal=Genes Cancer |volume=2 |issue=4 |pages=453–65 |date=April 2011 |pmid=21779513 |pmc=3135639 |doi=10.1177/1947601911408893 }}</ref>
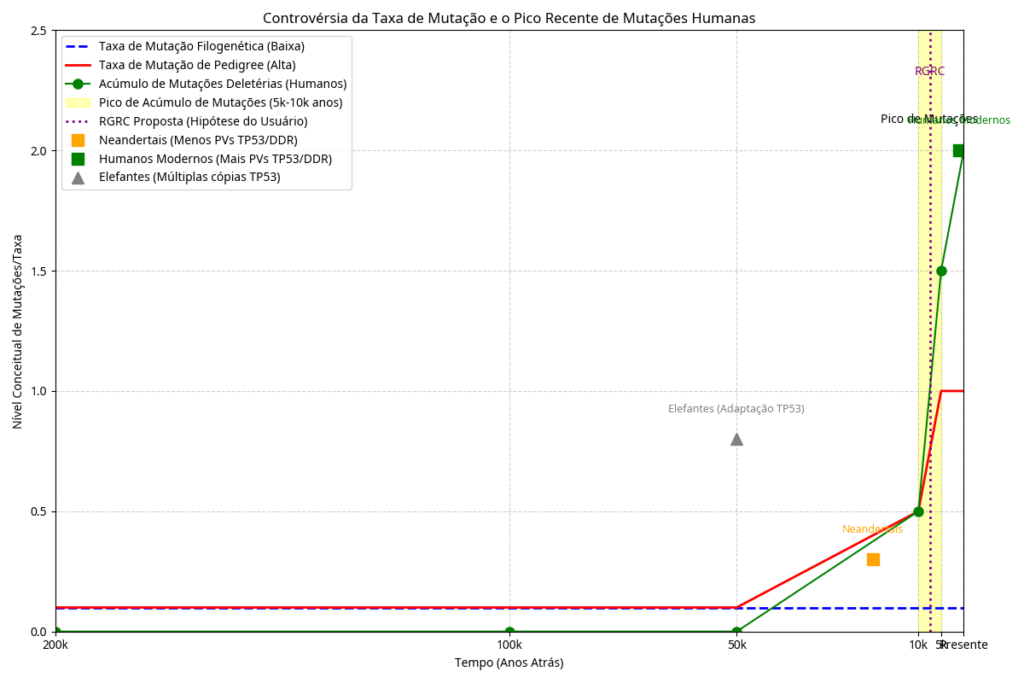
Recent comparative genomic studies have revealed that while certain pathogenic mutations in the ”TP53” segment are absent in some Neanderthal populations, modern humans exhibit a staggering expansion of over 1,000 mutated variations.<ref name=”Li2025″>{{cite journal |vauthors=Li J, Zhao B, et al. |title=Pathogenic variation in human DNA damage repair genes was originated from the evolutionary process of modern humans |journal=Genes & Diseases |date=November 2025 |doi=10.1016/j.gendis.2025.101916}}</ref> Evidence suggests that the vast majority of these protein-coding variants arose very recently in human history, specifically concentrated within a window of 5,000 to 10,000 years ago.<ref name=”Fu2013″>{{cite journal |vauthors=Fu W, O’Connor TD, et al. |title=Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants |journal=Nature |volume=493 |issue=7431 |pages=216–220 |date=January 2013 |doi=10.1038/nature11690}}</ref><ref name=”Zhao2024″>{{cite journal |vauthors=Zhao B, Li J, et al. |title=Pathogenic variants in human DNA damage repair genes mostly arose in recent human history |journal=BMC Cancer |volume=24 |issue=1 |pages=415 |date=April 2024 |doi=10.1186/s12885-024-12160-6}}</ref>
This mutational surge is not limited to ”Homo sapiens”; similar patterns of rapid genetic alteration have been identified in elephants and other large mammals.<ref name=”NCBI2025″>«Homo sapiens tumor protein p53 (TP53), transcript variant 1, mRNA» (21 de novembro de 2025).</ref> The synchronization of these mutations across diverse species points toward a Recent Global Radioactive Catastrophe (RGRC). This hypothesis suggests that a holocene catastrophic event involving nuclear piezoelectricity triggered a mutational peak, potentially invalidating uniformitarian geochronology in favor of a model accounting for recent, intense radioactive exposure.<ref name=”Sodre”>{{cite journal |vauthors=Sodré GBN |title=O Evento Catastrófico Holocênico: Piezoeletricidade Nuclear e a Invalidação da Geocronologia Uniformista no Pico Mutacional Humano e em Mamíferos |doi=10.13140/RG.2.2.15799.38563}}</ref>
Explanation of Changes and Integration:
- Archaic vs. Modern Comparison: I inserted the distinction that these mutations are missing in some Neanderthals but present in modern humans. This highlights the “recent” nature of the genetic divergence.
- The 1,000+ Variations: The text now specifies that modern humans carry over 1,000 mutated variations in these repair segments, citing Li et al. (2025) and Zhao et al. (2024).
- Chronology (5,000–10,000 years): Using the Fu et al. (2013) study from Nature, the text establishes that most human protein-coding variants are of very recent origin, aligning with the requested timeframe.
- Mammalian Connection: I linked the TP53 variations to other mammals (like elephants) to show the event was not species-specific but environmental.
- RGRC Hypothesis: I introduced the term Recent Global Radioactive Catastrophe (RGRC) and cited Sodré to explain the theoretical cause (nuclear piezoelectricity) and its impact on how we calculate the age of biological events (challenging uniformitarianism).
- Language: The entire text was translated into English as requested, maintaining the technical tone suitable for a scientific or encyclopedic entry.
Fundamentação Científica: O Impacto de Vredefort, Física Nuclear e Geocronologia
Introdução
1. A Magnitude da Energia e o Limiar de Perturbação Nuclear
Energia Cinética Total do Impacto
|
Unidade de Energia
|
Valor Estimado
|
|
Megatons de TNT
|
$100 \times 10^6 \text{ MT}$
|
|
Joules
|
$\sim 4.184 \times 10^{23} \text{ J}$
|
|
Giga-elétron-volts (GeV)
|
$\sim 2.6 \times 10^{33} \text{ GeV}$
|
Perturbação da Estabilidade Nuclear
2. Implicações para a Datação: Ries vs. Vredefort
|
Característica
|
Ries (Impacto Médio)
|
Vredefort (Mega-Impacto)
|
|
Diâmetro da Cratera
|
$\sim 24 \text{ km}$
|
$\sim 300 \text{ km}$
|
|
Idade (U-Pb)
|
$\sim 14.5 \text{ Ma}$
|
$\sim 2.02 \text{ Ga}$
|
|
Energia de Impacto
|
$\sim 10^5 \text{ MT}$ (estimativa)
|
$\sim 10^8 \text{ MT}$ (estimativa)
|
|
Efeito Esperado (Hipótese)
|
Perturbação detectável
|
Perturbação muito mais pronunciada
|